+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Initial DNA-lesion (Cy5) binding by XPC and TFIIH | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Protein-DNA complex / DNA BINDING PROTEIN-DNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationXPC complex / 9+2 motile cilium / MMXD complex / core TFIIH complex portion of holo TFIIH complex / photoreceptor connecting cilium / Cytosolic iron-sulfur cluster assembly / DNA damage sensor activity / central nervous system myelin formation / transcription export complex 2 / heterotrimeric G-protein binding ...XPC complex / 9+2 motile cilium / MMXD complex / core TFIIH complex portion of holo TFIIH complex / photoreceptor connecting cilium / Cytosolic iron-sulfur cluster assembly / DNA damage sensor activity / central nervous system myelin formation / transcription export complex 2 / heterotrimeric G-protein binding / regulation of proteasomal ubiquitin-dependent protein catabolic process / positive regulation of mitotic recombination / hair cell differentiation / hair follicle maturation / nucleotide-excision repair factor 3 complex / nucleotide-excision repair, preincision complex assembly / nuclear pore nuclear basket / CAK-ERCC2 complex / embryonic cleavage / DNA 5'-3' helicase / UV protection / regulation of cyclin-dependent protein serine/threonine kinase activity / transcription factor TFIIH core complex / transcription factor TFIIH holo complex / G protein-coupled receptor internalization / nuclear thyroid hormone receptor binding / cellular response to interleukin-7 / transcription preinitiation complex / RNA Polymerase I Transcription Termination / DNA 3'-5' helicase / RNA polymerase II general transcription initiation factor activity / regulation of mitotic cell cycle phase transition / transcription factor TFIID complex / erythrocyte maturation / 3'-5' DNA helicase activity / spinal cord development / hematopoietic stem cell proliferation / RNA Pol II CTD phosphorylation and interaction with CE during HIV infection / RNA Pol II CTD phosphorylation and interaction with CE / proteasome binding / HIV Transcription Initiation / RNA Polymerase II HIV Promoter Escape / Transcription of the HIV genome / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / bone mineralization / mRNA Capping / centriole replication / ATPase activator activity / DNA topological change / intrinsic apoptotic signaling pathway by p53 class mediator / RNA Polymerase I Transcription Initiation / polyubiquitin modification-dependent protein binding / glial cell projection / embryonic organ development / hematopoietic stem cell differentiation / mRNA transport / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / transcription elongation by RNA polymerase I / SUMOylation of DNA damage response and repair proteins / Formation of HIV elongation complex in the absence of HIV Tat / response to UV / transcription by RNA polymerase I / RNA Polymerase II Transcription Elongation / Formation of RNA Pol II elongation complex / hormone-mediated signaling pathway / transcription-coupled nucleotide-excision repair / extracellular matrix organization / RNA Polymerase II Pre-transcription Events / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / centriole / Recruitment of mitotic centrosome proteins and complexes / insulin-like growth factor receptor signaling pathway / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / DNA helicase activity / proteasome complex / AURKA Activation by TPX2 / ubiquitin binding / Josephin domain DUBs / regulation of cytokinesis / post-embryonic development / determination of adult lifespan / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / TP53 Regulates Transcription of DNA Repair Genes / nucleotide-excision repair / chromosome segregation / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / promoter-specific chromatin binding / RNA Polymerase I Promoter Escape / cellular response to gamma radiation / NoRC negatively regulates rRNA expression Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||
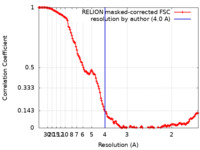
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | ||||||||||||
 Authors Authors | Kim J / Yang W | ||||||||||||
| Funding support |  United States, United States,  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Lesion recognition by XPC, TFIIH and XPA in DNA excision repair. Authors: Jinseok Kim / Chia-Lung Li / Xuemin Chen / Yanxiang Cui / Filip M Golebiowski / Huaibin Wang / Fumio Hanaoka / Kaoru Sugasawa / Wei Yang /     Abstract: Nucleotide excision repair removes DNA lesions caused by ultraviolet light, cisplatin-like compounds and bulky adducts. After initial recognition by XPC in global genome repair or a stalled RNA ...Nucleotide excision repair removes DNA lesions caused by ultraviolet light, cisplatin-like compounds and bulky adducts. After initial recognition by XPC in global genome repair or a stalled RNA polymerase in transcription-coupled repair, damaged DNA is transferred to the seven-subunit TFIIH core complex (Core7) for verification and dual incisions by the XPF and XPG nucleases. Structures capturing lesion recognition by the yeast XPC homologue Rad4 and TFIIH in transcription initiation or DNA repair have been separately reported. How two different lesion recognition pathways converge and how the XPB and XPD helicases of Core7 move the DNA lesion for verification are unclear. Here we report on structures revealing DNA lesion recognition by human XPC and DNA lesion hand-off from XPC to Core7 and XPA. XPA, which binds between XPB and XPD, kinks the DNA duplex and shifts XPC and the DNA lesion by nearly a helical turn relative to Core7. The DNA lesion is thus positioned outside of Core7, as would occur with RNA polymerase. XPB and XPD, which track the lesion-containing strand but translocate DNA in opposite directions, push and pull the lesion-containing strand into XPD for verification. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27996.map.gz emd_27996.map.gz | 17.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27996-v30.xml emd-27996-v30.xml emd-27996.xml emd-27996.xml | 36.8 KB 36.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27996_fsc.xml emd_27996_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27996.png emd_27996.png | 96.6 KB | ||
| Filedesc metadata |  emd-27996.cif.gz emd-27996.cif.gz | 10.5 KB | ||
| Others |  emd_27996_half_map_1.map.gz emd_27996_half_map_1.map.gz emd_27996_half_map_2.map.gz emd_27996_half_map_2.map.gz | 192.9 MB 192.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27996 http://ftp.pdbj.org/pub/emdb/structures/EMD-27996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27996 | HTTPS FTP |
-Related structure data
| Related structure data |  8ebsMC  8ebtC  8ebuC  8ebvC  8ebwC  8ebxC  8ebyC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27996.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27996.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27996_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27996_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : bulky DNA lesion recognition complex
+Supramolecule #1: bulky DNA lesion recognition complex
+Macromolecule #1: TFIIH basal transcription factor complex helicase XPB subunit
+Macromolecule #2: General transcription and DNA repair factor IIH helicase subunit XPD
+Macromolecule #3: General transcription factor IIH subunit 1
+Macromolecule #4: General transcription factor IIH subunit 4, p52
+Macromolecule #5: General transcription factor IIH subunit 2
+Macromolecule #6: General transcription factor IIH subunit 3
+Macromolecule #7: General transcription factor IIH subunit 5
+Macromolecule #8: Xeroderma pigmentosum, complementation group C, isoform CRA_a
+Macromolecule #9: UV excision repair protein RAD23 homolog B
+Macromolecule #10: Centrin-2
+Macromolecule #11: DNA (Cy5)
+Macromolecule #12: DNA
+Macromolecule #13: IRON/SULFUR CLUSTER
+Macromolecule #14: ZINC ION
+Macromolecule #15: CALCIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 6243 / Average exposure time: 2.5 sec. / Average electron dose: 54.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.6 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)