+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pseudomonas phage E217 contracted sheath | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pseudomonas / phage / E217 / capsid / decorating proteins / VIRUS | |||||||||
| Function / homology | Protein of unknown function DUF3383 / Protein of unknown function (DUF3383) / Tail sheath protein Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) | |||||||||
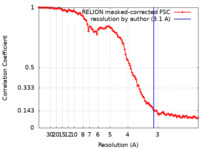
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Li F / Cingolani G / Hou C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217. Authors: Fenglin Li / Chun-Feng David Hou / Ravi K Lokareddy / Ruoyu Yang / Francesca Forti / Federica Briani / Gino Cingolani /   Abstract: E217 is a Pseudomonas phage used in an experimental cocktail to eradicate cystic fibrosis-associated Pseudomonas aeruginosa. Here, we describe the structure of the whole E217 virion before and after ...E217 is a Pseudomonas phage used in an experimental cocktail to eradicate cystic fibrosis-associated Pseudomonas aeruginosa. Here, we describe the structure of the whole E217 virion before and after DNA ejection at 3.1 Å and 4.5 Å resolution, respectively, determined using cryogenic electron microscopy (cryo-EM). We identify and build de novo structures for 19 unique E217 gene products, resolve the tail genome-ejection machine in both extended and contracted states, and decipher the complete architecture of the baseplate formed by 66 polypeptide chains. We also determine that E217 recognizes the host O-antigen as a receptor, and we resolve the N-terminal portion of the O-antigen-binding tail fiber. We propose that E217 design principles presented in this paper are conserved across PB1-like Myoviridae phages of the Pbunavirus genus that encode a ~1.4 MDa baseplate, dramatically smaller than the coliphage T4. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29486.map.gz emd_29486.map.gz | 79.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29486-v30.xml emd-29486-v30.xml emd-29486.xml emd-29486.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29486_fsc.xml emd_29486_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29486.png emd_29486.png | 191.4 KB | ||
| Filedesc metadata |  emd-29486.cif.gz emd-29486.cif.gz | 5.1 KB | ||
| Others |  emd_29486_half_map_1.map.gz emd_29486_half_map_1.map.gz emd_29486_half_map_2.map.gz emd_29486_half_map_2.map.gz | 79.2 MB 79.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29486 http://ftp.pdbj.org/pub/emdb/structures/EMD-29486 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29486 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29486 | HTTPS FTP |
-Validation report
| Summary document |  emd_29486_validation.pdf.gz emd_29486_validation.pdf.gz | 1.4 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29486_full_validation.pdf.gz emd_29486_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  emd_29486_validation.xml.gz emd_29486_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_29486_validation.cif.gz emd_29486_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29486 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29486 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29486 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29486 | HTTPS FTP |
-Related structure data
| Related structure data |  8fvgMC  8envC  8eonC  8frsC  8fuvC  8fvhC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29486.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29486.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_29486_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29486_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pseudomonas phage vB_PaeM_E217
| Entire | Name:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage vB_PaeM_E217
| Supramolecule | Name: Pseudomonas phage vB_PaeM_E217 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2034346 / Sci species name: Pseudomonas phage vB_PaeM_E217 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: Sheath protein gp31
| Macromolecule | Name: Sheath protein gp31 / type: protein_or_peptide / ID: 1 / Details: contracted state / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage vB_PaeM_E217 (virus) Pseudomonas phage vB_PaeM_E217 (virus) |
| Molecular weight | Theoretical: 53.680723 KDa |
| Sequence | String: MISQSRYIRI ISGVGAGAPV AGRKLILRVM TTNNVIPPGI VIEFDNANAV LSYFGAQSEE YQRAAAYFKF ISKSVNSPSS ISFARWVNT AIAPMVVGDN LPKTIADFAG FSAGVLTIMV GAAEQNITAI DTSAATSMDN VASIIQTEIR KNADPQLAQA T VTWNQNTN ...String: MISQSRYIRI ISGVGAGAPV AGRKLILRVM TTNNVIPPGI VIEFDNANAV LSYFGAQSEE YQRAAAYFKF ISKSVNSPSS ISFARWVNT AIAPMVVGDN LPKTIADFAG FSAGVLTIMV GAAEQNITAI DTSAATSMDN VASIIQTEIR KNADPQLAQA T VTWNQNTN QFTLVGATIG TGVLAVAKSA DPQDMSTALG WSTSNVVNVA GQSADLPDAA VAKSTNVSNN FGSFLFAGAP LD NDQIKAV SAWNAAQNNQ FIYTVATSLA NLGTLFTLVN GNAGTALNVL SATAANDFVE QCPSEILAAT NYDEPGASQN YMY YQFPGR NITVSDDTVA NTVDKSRGNY IGVTQANGQQ LAFYQRGILC GGPTDAVDMN VYANEIWLKS AIAQALLDLF LNVN AVPAS STGEAMTLAV LQPVLDKATA NGTFTYGKEI SAVQQQYITQ VTGDRRAWRQ VQTLGYWINI TFSSYTNSNT GLTEW KANY TLIYSKGDAI RFVEGSDVMI UniProtKB: Tail sheath protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)