[English] 日本語
 Yorodumi
Yorodumi- EMDB-29447: Sec39:Use1:Sec20:Tip20 Local Refinement of the Dsl1:Qb:Qc Complex #2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sec39:Use1:Sec20:Tip20 Local Refinement of the Dsl1:Qb:Qc Complex #2 | |||||||||
 Map data Map data | Sharpened Map, Local Refinement #1, of the Dsl1:Qb:Qc complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tether / SNARE / Complex / TRANSPORT PROTEIN | |||||||||
| Biological species |  | |||||||||
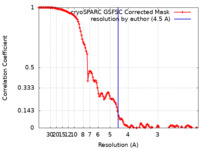
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | DAmico KA / Jeffrey PD / Hughson FM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of a membrane tethering complex incorporating multiple SNAREs. Authors: Kevin A DAmico / Abigail E Stanton / Jaden D Shirkey / Sophie M Travis / Philip D Jeffrey / Frederick M Hughson /  Abstract: Most membrane fusion reactions in eukaryotic cells are mediated by multisubunit tethering complexes (MTCs) and SNARE proteins. MTCs are much larger than SNAREs and are thought to mediate the initial ...Most membrane fusion reactions in eukaryotic cells are mediated by multisubunit tethering complexes (MTCs) and SNARE proteins. MTCs are much larger than SNAREs and are thought to mediate the initial attachment of two membranes. Complementary SNAREs then form membrane-bridging complexes whose assembly draws the membranes together for fusion. Here we present a cryo-electron microscopy structure of the simplest known MTC, the 255-kDa Dsl1 complex of Saccharomyces cerevisiae, bound to the two SNAREs that anchor it to the endoplasmic reticulum. N-terminal domains of the SNAREs form an integral part of the structure, stabilizing a Dsl1 complex configuration with unexpected similarities to the 850-kDa exocyst MTC. The structure of the SNARE-anchored Dsl1 complex and its comparison with exocyst reveal what are likely to be common principles underlying MTC function. Our structure also implies that tethers and SNAREs can work together as a single integrated machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29447.map.gz emd_29447.map.gz | 230.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29447-v30.xml emd-29447-v30.xml emd-29447.xml emd-29447.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29447_fsc.xml emd_29447_fsc.xml | 13.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_29447.png emd_29447.png | 73.7 KB | ||
| Masks |  emd_29447_msk_1.map emd_29447_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29447.cif.gz emd-29447.cif.gz | 6.8 KB | ||
| Others |  emd_29447_additional_1.map.gz emd_29447_additional_1.map.gz emd_29447_half_map_1.map.gz emd_29447_half_map_1.map.gz emd_29447_half_map_2.map.gz emd_29447_half_map_2.map.gz | 121.4 MB 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29447 http://ftp.pdbj.org/pub/emdb/structures/EMD-29447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29447 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29447 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29447.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29447.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map, Local Refinement #1, of the Dsl1:Qb:Qc complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
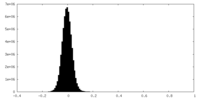
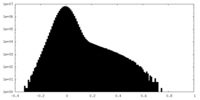
-Mask #1
| File |  emd_29447_msk_1.map emd_29447_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
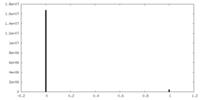
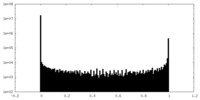
| Density Histograms |
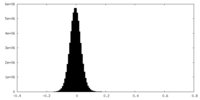
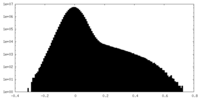
-Additional map: Unsharpened Map, Local Refinement #1, of the Dsl1:Qb:Qc complex
| File | emd_29447_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Map, Local Refinement #1, of the Dsl1:Qb:Qc complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
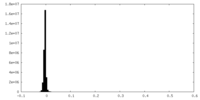
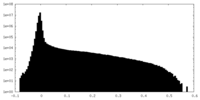
| Density Histograms |
-Half map: Half-Map A, Local Refinement #1, of the Dsl1:Qb:Qc complex
| File | emd_29447_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-Map A, Local Refinement #1, of the Dsl1:Qb:Qc complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-Map B, Local Refinement #1, of the Dsl1:Qb:Qc complex
| File | emd_29447_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-Map B, Local Refinement #1, of the Dsl1:Qb:Qc complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dsl1 complex bound to SNARE proteins Sec20 and Use1
| Entire | Name: Dsl1 complex bound to SNARE proteins Sec20 and Use1 |
|---|---|
| Components |
|
-Supramolecule #1: Dsl1 complex bound to SNARE proteins Sec20 and Use1
| Supramolecule | Name: Dsl1 complex bound to SNARE proteins Sec20 and Use1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 312.34826 KDa |
-Macromolecule #1: Sec39
| Macromolecule | Name: Sec39 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MLEEQLYLLA CIFASRADTR NIKKLSTRLG SQSKYLEILC VLWPELDDPK NLLFLRELEE EVQSPEGEET TDEDVIVELL ESDSSLIPLI ESDTTTRSNR YHELQEFISK KLNNKTLENF EEWLRERILI CNEMIPETPL LYSVLWETAK SKVLSTKFIG WVEGVLKPLD ...String: MLEEQLYLLA CIFASRADTR NIKKLSTRLG SQSKYLEILC VLWPELDDPK NLLFLRELEE EVQSPEGEET TDEDVIVELL ESDSSLIPLI ESDTTTRSNR YHELQEFISK KLNNKTLENF EEWLRERILI CNEMIPETPL LYSVLWETAK SKVLSTKFIG WVEGVLKPLD HLNKRLHLIF KINEWEKMPD SELFKIIFDG VEDMQGYIGI ADVIEDELAP TLSYGKKWET FITEFFNKQQ FSLKSDTNYQ LFIKLYYSLE KGVKDNSEAS RKLQSNVVDI LFHNSENLFN LSSLTHKLDE LWSILSGFPD EITIEEQKTI TALEMKQFME FFIKCSTKFS FKEIFAITQE EESAQLAHFS SLCHEEFNKA NEISSFLQAM YETVLDISKD DKIFTRISMD EKLYSILEIL LQMNEFAYIE AIIERFDYSN NTQIYELLVK FFWHFFNNAS NGLRKEPEMK KASQTLQIIQ KHMSQRAGTN LTKLEVLLEI SDKLSHYSIN LNKSHNGARD TAFKPSNILE YRDCPLDIIS NLLELNPRLY KDLPTTKSLL FGIYDSLSIN REGQTGKVEV DLMVLHIDYA LVNLDFGTAY ELGKQVFEIC QEAGQHMMKA LGDEHWLTFY QMGKFVDPNW VDNEIPTEII VLQMSILGRL LEVCPLEEVE IVTSQWSTLE LELSARDLVK DKYALDGQND NKSKVGGIAR EIFHNVTNF |
| Recombinant expression | Organism:  |
-Macromolecule #2: Use1
| Macromolecule | Name: Use1 / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MAETSNDPFL SYVLSSKQLT NLNRLRRKAV TKQLGSSDDN KVSEEFLRYQ HTYQREAFEY LQTKHDAHKI MESQYEQYQS SSKTRRYSID LDSVDAVDTE SQTEYPNEEF IDRNEDSEAV MELRKRLLGK GQNKGLGYET TKSVDRQIED QDTLQQDLIQ DMSKLVGSLK ...String: MAETSNDPFL SYVLSSKQLT NLNRLRRKAV TKQLGSSDDN KVSEEFLRYQ HTYQREAFEY LQTKHDAHKI MESQYEQYQS SSKTRRYSID LDSVDAVDTE SQTEYPNEEF IDRNEDSEAV MELRKRLLGK GQNKGLGYET TKSVDRQIED QDTLQQDLIQ DMSKLVGSLK QGAVAFQSAL DEDKQVLGAA EIGIQVASQG LMDVSGKLRK YD |
-Macromolecule #3: Sec20
| Macromolecule | Name: Sec20 / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MVVTFLQDLE VLQDALLNNL QKLSAISRRK ESGESKHDNK DSFAAIANEH NDEEEEIEFE DLVNIIESKV SDFESVLKCS IVEMTYKYPE LKLQWEKSPR YDQCDKLHIV KLDKQMNEDI YAQLVEELDF VLQFVDWFYC YRLKVKEILR QHHKRDLAWN DEKRDRAIKF ...String: MVVTFLQDLE VLQDALLNNL QKLSAISRRK ESGESKHDNK DSFAAIANEH NDEEEEIEFE DLVNIIESKV SDFESVLKCS IVEMTYKYPE LKLQWEKSPR YDQCDKLHIV KLDKQMNEDI YAQLVEELDF VLQFVDWFYC YRLKVKEILR QHHKRDLAWN DEKRDRAIKF HAVDYDKLHQ GTSSSSSLTS TSMEKASTRE KLLSKTKQLT NNLVRGNQIL QSGILQSDLN LDELRAQTNS LTQIDDKYTQ FETVFKKTAD LVKVLENASH QEKRD |
-Macromolecule #4: Tip20
| Macromolecule | Name: Tip20 / type: other / ID: 4 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MNGIDDLLNI NDRIKQVQNE RNELASKLQN LKQSLASNDT EVALSEVIAQ DIIEVGASVE GLEQLRAKYG DLQILNKLEK VAVQQTQMQA GVDKLDSFER QLDELAEQPP DQFTLDDVKA LHSKLTSVFA TVPQINNIDS QYAAYNKLKS KVTGKYNDVI IQRLATNWSN ...String: MNGIDDLLNI NDRIKQVQNE RNELASKLQN LKQSLASNDT EVALSEVIAQ DIIEVGASVE GLEQLRAKYG DLQILNKLEK VAVQQTQMQA GVDKLDSFER QLDELAEQPP DQFTLDDVKA LHSKLTSVFA TVPQINNIDS QYAAYNKLKS KVTGKYNDVI IQRLATNWSN TFDQKLLEAQ WDTQKFASTS VGLVKCLREN STKLYQLSLL YLPLEEETQN GDSERPLSRS NNNQEPVLWN FKSLANNFNV RFTYHFHATS SSSKIETYFQ FLNDYLAENL YKCINIFHDD CNGLTKPVIH EQFINYVLQP IRDKVRSTLF QNDLKTLIVL ISQILATDKN LLNSFHYHGL GLVSLISDEV WEKWINYEVE MANRQFINIT KNPEDFPKSS QNFVKLINKI YDYLEPFYDL DFDLLVRYKL MTCSLIFMNL TSSYLDYILT VDSLNETRTK EQELYQTMAK LQHVNFVYRK IKSLSSNFIF IQLTDIVNST ESKKYNSLFQ NVENDYEKAM STDMQNSIVH RIQKLLKETL RNYFKISTWS TLEMSVDENI GPSSVPSAEL VNSINVLRRL INKLDSMDIP LAISLKVKNE LLNVIVNYFT ESILKLNKFN QNGLNQFLHD FKSLSSILSL PSHATNYKCM SLHELVKILK LKYDPNNQQF LNPEYIKTGN FTSLKEAYSI KYLKDTKIQD ALYRIIYGNI L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Buffer was made fresh from concentrated components and sterile filtered. NP40 was not present during protein purification but was an additive during the grid preparation. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 10 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | Sample was consistently in the thickest regions of ice only, often close to the edges of the carbon hole |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 5857 / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)