[English] 日本語
 Yorodumi
Yorodumi- EMDB-28204: CryoEM structure of the Dsl1 complex bound to SNAREs Sec20 and Use1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the Dsl1 complex bound to SNAREs Sec20 and Use1 | |||||||||
 Map data Map data | Half map B | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tether / SNARE / Complex / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationGolgi to ER transport vesicle membrane / vesicle fusion with endoplasmic reticulum / ER-dependent peroxisome organization / Dsl1/NZR complex / RZZ complex / regulation of ER to Golgi vesicle-mediated transport / COPI-dependent Golgi-to-ER retrograde traffic / SNARE complex / SNAP receptor activity / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum ...Golgi to ER transport vesicle membrane / vesicle fusion with endoplasmic reticulum / ER-dependent peroxisome organization / Dsl1/NZR complex / RZZ complex / regulation of ER to Golgi vesicle-mediated transport / COPI-dependent Golgi-to-ER retrograde traffic / SNARE complex / SNAP receptor activity / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / mitotic spindle assembly checkpoint signaling / cytoplasmic side of endoplasmic reticulum membrane / endoplasmic reticulum to Golgi vesicle-mediated transport / vesicle-mediated transport / autophagy / nuclear envelope / protein transport / endoplasmic reticulum membrane / endoplasmic reticulum / membrane / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | DAmico KA / Jeffrey PD / Hughson FM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure of a membrane tethering complex incorporating multiple SNAREs. Authors: Kevin A DAmico / Abigail E Stanton / Jaden D Shirkey / Sophie M Travis / Philip D Jeffrey / Frederick M Hughson /  Abstract: Most membrane fusion reactions in eukaryotic cells are mediated by multisubunit tethering complexes (MTCs) and SNARE proteins. MTCs are much larger than SNAREs and are thought to mediate the initial ...Most membrane fusion reactions in eukaryotic cells are mediated by multisubunit tethering complexes (MTCs) and SNARE proteins. MTCs are much larger than SNAREs and are thought to mediate the initial attachment of two membranes. Complementary SNAREs then form membrane-bridging complexes whose assembly draws the membranes together for fusion. Here we present a cryo-electron microscopy structure of the simplest known MTC, the 255-kDa Dsl1 complex of Saccharomyces cerevisiae, bound to the two SNAREs that anchor it to the endoplasmic reticulum. N-terminal domains of the SNAREs form an integral part of the structure, stabilizing a Dsl1 complex configuration with unexpected similarities to the 850-kDa exocyst MTC. The structure of the SNARE-anchored Dsl1 complex and its comparison with exocyst reveal what are likely to be common principles underlying MTC function. Our structure also implies that tethers and SNAREs can work together as a single integrated machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28204.map.gz emd_28204.map.gz | 200 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28204-v30.xml emd-28204-v30.xml emd-28204.xml emd-28204.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28204_fsc.xml emd_28204_fsc.xml emd_28204_fsc_2.xml emd_28204_fsc_2.xml emd_28204_fsc_3.xml emd_28204_fsc_3.xml emd_28204_fsc_4.xml emd_28204_fsc_4.xml | 13.4 KB 13.4 KB 13.4 KB 13.3 KB | Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_28204.png emd_28204.png | 57.4 KB | ||
| Filedesc metadata |  emd-28204.cif.gz emd-28204.cif.gz | 8.3 KB | ||
| Others |  emd_28204_half_map_1.map.gz emd_28204_half_map_1.map.gz emd_28204_half_map_2.map.gz emd_28204_half_map_2.map.gz | 198.5 MB 198.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28204 http://ftp.pdbj.org/pub/emdb/structures/EMD-28204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28204 | HTTPS FTP |
-Related structure data
| Related structure data |  8ekiMC  8ftuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28204.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28204.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.114 Å | ||||||||||||||||||||||||||||||||||||
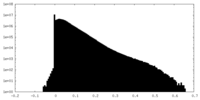
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_28204_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
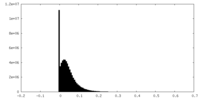
| Density Histograms |
-Half map: Half map A
| File | emd_28204_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
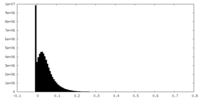
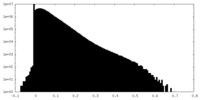
| Density Histograms |
- Sample components
Sample components
-Entire : Dsl1 complex bound to SNARE proteins Sec20 and Use1
| Entire | Name: Dsl1 complex bound to SNARE proteins Sec20 and Use1 |
|---|---|
| Components |
|
-Supramolecule #1: Dsl1 complex bound to SNARE proteins Sec20 and Use1
| Supramolecule | Name: Dsl1 complex bound to SNARE proteins Sec20 and Use1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 255.41261 KDa |
-Supramolecule #2: Dsl1 Complex
| Supramolecule | Name: Dsl1 Complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein transport protein SEC20
| Macromolecule | Name: Protein transport protein SEC20 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.244254 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVVTFLQDLE VLQDALLNNL QKLSAISRRK ESGESKHDNK DSFAAIANEH NDEEEEIEFE DLVNIIESKV SDFESVLKCS IVEMTYKYP ELKLQWEKSP RYDQCDKLHI VKLDKQMNED IYAQLVEELD FVLQFVDWFY CYRLKVKEIL RQHHKRDLAW N DEKRDRAI ...String: MVVTFLQDLE VLQDALLNNL QKLSAISRRK ESGESKHDNK DSFAAIANEH NDEEEEIEFE DLVNIIESKV SDFESVLKCS IVEMTYKYP ELKLQWEKSP RYDQCDKLHI VKLDKQMNED IYAQLVEELD FVLQFVDWFY CYRLKVKEIL RQHHKRDLAW N DEKRDRAI KFHAVDYDKL HQGTSSSSSL TSTSMEKAST REKLLSKTKQ LTNNLVRGNQ ILQSGILQSD LNLDELRAQT NS LTQIDDK YTQFETVFKK TADLVKVLEN ASHQEKRD UniProtKB: Protein transport protein SEC20 |
-Macromolecule #2: Protein transport protein USE1
| Macromolecule | Name: Protein transport protein USE1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.254781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAETSNDPFL SYVLSSKQLT NLNRLRRKAV TKQLGSSDDN KVSEEFLRYQ HTYQREAFEY LQTKHDAHKI MESQYEQYQS SSKTRRYSI DLDSVDAVDT ESQTEYPNEE FIDRNEDSEA VMELRKRLLG KGQNKGLGYE TTKSVDRQIE DQDTLQQDLI Q DMSKLVGS ...String: MAETSNDPFL SYVLSSKQLT NLNRLRRKAV TKQLGSSDDN KVSEEFLRYQ HTYQREAFEY LQTKHDAHKI MESQYEQYQS SSKTRRYSI DLDSVDAVDT ESQTEYPNEE FIDRNEDSEA VMELRKRLLG KGQNKGLGYE TTKSVDRQIE DQDTLQQDLI Q DMSKLVGS LKQGAVAFQS ALDEDKQVLG AAEIGIQVAS QGLMDVSGKL RKYD UniProtKB: Protein transport protein USE1 |
-Macromolecule #3: Protein transport protein TIP20
| Macromolecule | Name: Protein transport protein TIP20 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.253062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNGIDDLLNI NDRIKQVQNE RNELASKLQN LKQSLASNDT EVALSEVIAQ DIIEVGASVE GLEQLRAKYG DLQILNKLEK VAVQQTQMQ AGVDKLDSFE RQLDELAEQP PDQFTLDDVK ALHSKLTSVF ATVPQINNID SQYAAYNKLK SKVTGKYNDV I IQRLATNW ...String: MNGIDDLLNI NDRIKQVQNE RNELASKLQN LKQSLASNDT EVALSEVIAQ DIIEVGASVE GLEQLRAKYG DLQILNKLEK VAVQQTQMQ AGVDKLDSFE RQLDELAEQP PDQFTLDDVK ALHSKLTSVF ATVPQINNID SQYAAYNKLK SKVTGKYNDV I IQRLATNW SNTFDQKLLE AQWDTQKFAS TSVGLVKCLR ENSTKLYQLS LLYLPLEEET QNGDSERPLS RSNNNQEPVL WN FKSLANN FNVRFTYHFH ATSSSSKIET YFQFLNDYLA ENLYKCINIF HDDCNGLTKP VIHEQFINYV LQPIRDKVRS TLF QNDLKT LIVLISQILA TDKNLLNSFH YHGLGLVSLI SDEVWEKWIN YEVEMANRQF INITKNPEDF PKSSQNFVKL INKI YDYLE PFYDLDFDLL VRYKLMTCSL IFMNLTSSYL DYILTVDSLN ETRTKEQELY QTMAKLQHVN FVYRKIKSLS SNFIF IQLT DIVNSTESKK YNSLFQNVEN DYEKAMSTDM QNSIVHRIQK LLKETLRNYF KISTWSTLEM SVDENIGPSS VPSAEL VNS INVLRRLINK LDSMDIPLAI SLKVKNELLN VIVNYFTESI LKLNKFNQNG LNQFLHDFKS LSSILSLPSH ATNYKCM SL HELVKILKLK YDPNNQQFLN PEYIKTGNFT SLKEAYSIKY LKDTKIQDAL YRIIYGNIL UniProtKB: Protein transport protein TIP20 |
-Macromolecule #4: Protein transport protein SEC39
| Macromolecule | Name: Protein transport protein SEC39 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 82.483797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLEEQLYLLA CIFASRADTR NIKKLSTRLG SQSKYLEILC VLWPELDDPK NLLFLRELEE EVQSPEGEET TDEDVIVELL ESDSSLIPL IESDTTTRSN RYHELQEFIS KKLNNKTLEN FEEWLRERIL ICNEMIPETP LLYSVLWETA KSKVLSTKFI G WVEGVLKP ...String: MLEEQLYLLA CIFASRADTR NIKKLSTRLG SQSKYLEILC VLWPELDDPK NLLFLRELEE EVQSPEGEET TDEDVIVELL ESDSSLIPL IESDTTTRSN RYHELQEFIS KKLNNKTLEN FEEWLRERIL ICNEMIPETP LLYSVLWETA KSKVLSTKFI G WVEGVLKP LDHLNKRLHL IFKINEWEKM PDSELFKIIF DGVEDMQGYI GIADVIEDEL APTLSYGKKW ETFITEFFNK QQ FSLKSDT NYQLFIKLYY SLEKGVKDNS EASRKLQSNV VDILFHNSEN LFNLSSLTHK LDELWSILSG FPDEITIEEQ KTI TALEMK QFMEFFIKCS TKFSFKEIFA ITQEEESAQL AHFSSLCHEE FNKANEISSF LQAMYETVLD ISKDDKIFTR ISMD EKLYS ILEILLQMNE FAYIEAIIER FDYSNNTQIY ELLVKFFWHF FNNASNGLRK EPEMKKASQT LQIIQKHMSQ RAGTN LTKL EVLLEISDKL SHYSINLNKS HNGARDTAFK PSNILEYRDC PLDIISNLLE LNPRLYKDLP TTKSLLFGIY DSLSIN REG QTGKVEVDLM VLHIDYALVN LDFGTAYELG KQVFEICQEA GQHMMKALGD EHWLTFYQMG KFVDPNWVDN EIPTEII VL QMSILGRLLE VCPLEEVEIV TSQWSTLELE LSARDLVKDK YALDGQNDNK SKVGGIAREI FHNVTNF UniProtKB: Protein transport protein SEC39 |
-Macromolecule #5: Protein transport protein DSL1
| Macromolecule | Name: Protein transport protein DSL1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 91.445133 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKHHHHHHHG AAGTSLYKKA GENLYFQGSM ESLFPNKGEI IRELLKDPLI LKNDSKRSNG SELELDSSDL LQREAILANE LNILDNLKT FLNLIKEVKT NLNILELENC YYSLQSLRKK MRNNAAYLKQ SFNFQQSIST YVDTLHLELV STLYKILTNG F WKITENSI ...String: MKHHHHHHHG AAGTSLYKKA GENLYFQGSM ESLFPNKGEI IRELLKDPLI LKNDSKRSNG SELELDSSDL LQREAILANE LNILDNLKT FLNLIKEVKT NLNILELENC YYSLQSLRKK MRNNAAYLKQ SFNFQQSIST YVDTLHLELV STLYKILTNG F WKITENSI QFTPTVEWGK DKVHIEYDTF MDFVAQQYFP KGSLDNQAWF ILDMTSADSQ EQVRAKLNTI MKEYMNLSRI VS MIKNSIF ISGKEISYEN EKNILVFSKS SSHGQHCVST VLTSFEAVCD FMLDGLAFRD RKTLSYELGP LFNTEFTKFV KNN ASIILE SLDSPLKNLV SVINNKLTRL VAKSEVTNWT HSGKEIQDLL MNKQLYYNLL LDKVLESHIS EIRSIFEDPK KSWQ NLEVV ELTTSNTNTM SEKIGKNDSD VQNEKELHNA VSKDDDWNWE VEDDDADAWG DEIDVNIDDE EEKTNQEKEK EPEEE ENAW DEAWAIDENI DDASLENGKE HLKAHDVGSL DKDHIEVTQL PKLFLAISQN FKSSFADSHV DEQYFAYKYN LLQTSY MAM CTANFSHNWC QLYVDMRYLI ERDEKLYRIK ELTRNLLETK LNMKYRIVCQ LIRHQLTEFR ENERNPSWDA TIEKLLP YI LKEIVRPLQK IRGEEGSRYL LSFLNFLYND CVTKEILKWQ IISEVNSENL GELVSLLVNN TDIQLLAKEP SYKKMREK F ATMGKFLPLH LKEIMEMFYN GDFYLFATDE LIQWIELLFA DTPLRRNAID DIYEIRGTAL DD UniProtKB: Protein transport protein DSL1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Buffer was made fresh from concentrated components and sterile filtered. NP40 was not present during protein purification but was an additive during the grid preparation. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 10 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Force=0 Wait Time=0 Blot Time=6s Drain Time=0. | |||||||||||||||
| Details | Sample was consistently in the thickest regions of ice only, often close to the edges of the carbon hole |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 5857 / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)