[English] 日本語
 Yorodumi
Yorodumi- EMDB-29365: Co-structure of the Respiratory Syncytial Virus RNA-dependent RNA... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Co-structure of the Respiratory Syncytial Virus RNA-dependent RNA polymerase with MRK-1 | |||||||||
 Map data Map data | sharpened map used for model building and real space refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-BINDING PROTEIN / RSV / RDRP / RNA-DEPENDENT RNA POLYMERASE / PRNTASE / POLYRIBONUCLEOTIDYL TRANSFERASE / RNA CAPPING / VIRAL REPLICATION / VIRAL PROTEIN / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationRespiratory syncytial virus genome transcription / NNS virus cap methyltransferase / Translation of respiratory syncytial virus mRNAs / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / Respiratory syncytial virus genome replication / RSV-host interactions / Assembly and release of respiratory syncytial virus (RSV) virions / Maturation of hRSV A proteins / Respiratory syncytial virus (RSV) attachment and entry ...Respiratory syncytial virus genome transcription / NNS virus cap methyltransferase / Translation of respiratory syncytial virus mRNAs / GDP polyribonucleotidyltransferase / negative stranded viral RNA replication / Respiratory syncytial virus genome replication / RSV-host interactions / Assembly and release of respiratory syncytial virus (RSV) virions / Maturation of hRSV A proteins / Respiratory syncytial virus (RSV) attachment and entry / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / viral life cycle / virion component / symbiont-mediated suppression of host NF-kappaB cascade / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / GTPase activity / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Human orthopneumovirus / Human orthopneumovirus /  Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.52 Å | |||||||||
 Authors Authors | Fischmann TO | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Conserved allosteric inhibitory site on the respiratory syncytial virus and human metapneumovirus RNA-dependent RNA polymerases. Authors: Victoria A Kleiner / Thierry O Fischmann / John A Howe / Douglas C Beshore / Michael J Eddins / Yan Hou / Todd Mayhood / Daniel Klein / Debbie D Nahas / Bob J Lucas / He Xi / Edward Murray / ...Authors: Victoria A Kleiner / Thierry O Fischmann / John A Howe / Douglas C Beshore / Michael J Eddins / Yan Hou / Todd Mayhood / Daniel Klein / Debbie D Nahas / Bob J Lucas / He Xi / Edward Murray / Daphne Y Ma / Krista Getty / Rachel Fearns /  Abstract: Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) are related RNA viruses responsible for severe respiratory infections and resulting disease in infants, elderly, and ...Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) are related RNA viruses responsible for severe respiratory infections and resulting disease in infants, elderly, and immunocompromised adults. Therapeutic small molecule inhibitors that bind to the RSV polymerase and inhibit viral replication are being developed, but their binding sites and molecular mechanisms of action remain largely unknown. Here we report a conserved allosteric inhibitory site identified on the L polymerase proteins of RSV and HMPV that can be targeted by a dual-specificity, non-nucleoside inhibitor, termed MRK-1. Cryo-EM structures of the inhibitor in complexes with truncated RSV and full-length HMPV polymerase proteins provide a structural understanding of how MRK-1 is active against both viruses. Functional analyses indicate that MRK-1 inhibits conformational changes necessary for the polymerase to engage in RNA synthesis initiation and to transition into an elongation mode. Competition studies reveal that the MRK-1 binding pocket is distinct from that of a capping inhibitor with an overlapping resistance profile, suggesting that the polymerase conformation bound by MRK-1 may be distinct from that involved in mRNA capping. These findings should facilitate optimization of dual RSV and HMPV replication inhibitors and provide insights into the molecular mechanisms underlying their polymerase activities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29365.map.gz emd_29365.map.gz | 28.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29365-v30.xml emd-29365-v30.xml emd-29365.xml emd-29365.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29365_fsc.xml emd_29365_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29365.png emd_29365.png | 130.1 KB | ||
| Filedesc metadata |  emd-29365.cif.gz emd-29365.cif.gz | 7.5 KB | ||
| Others |  emd_29365_half_map_1.map.gz emd_29365_half_map_1.map.gz emd_29365_half_map_2.map.gz emd_29365_half_map_2.map.gz | 28.4 MB 28.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29365 http://ftp.pdbj.org/pub/emdb/structures/EMD-29365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29365 | HTTPS FTP |
-Validation report
| Summary document |  emd_29365_validation.pdf.gz emd_29365_validation.pdf.gz | 814.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29365_full_validation.pdf.gz emd_29365_full_validation.pdf.gz | 814.3 KB | Display | |
| Data in XML |  emd_29365_validation.xml.gz emd_29365_validation.xml.gz | 14.2 KB | Display | |
| Data in CIF |  emd_29365_validation.cif.gz emd_29365_validation.cif.gz | 18.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29365 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29365 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29365 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29365 | HTTPS FTP |
-Related structure data
| Related structure data |  8fpiMC  8fpjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29365.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29365.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map used for model building and real space refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_29365_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
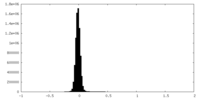
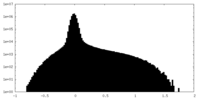
| Density Histograms |
-Half map: half map
| File | emd_29365_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
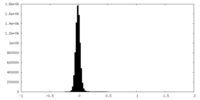
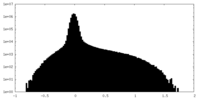
| Density Histograms |
- Sample components
Sample components
-Entire : RESPIRATORY SYNCYTIAL VIRUS POLYMERASE (L) PROTEIN BOUND BY THE T...
| Entire | Name: RESPIRATORY SYNCYTIAL VIRUS POLYMERASE (L) PROTEIN BOUND BY THE TETRAMERIC PHOSPHOPROTEIN (P) AND COMPLEXED WITH MRK-1 |
|---|---|
| Components |
|
-Supramolecule #1: RESPIRATORY SYNCYTIAL VIRUS POLYMERASE (L) PROTEIN BOUND BY THE T...
| Supramolecule | Name: RESPIRATORY SYNCYTIAL VIRUS POLYMERASE (L) PROTEIN BOUND BY THE TETRAMERIC PHOSPHOPROTEIN (P) AND COMPLEXED WITH MRK-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Human orthopneumovirus Human orthopneumovirus |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 173.267922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSDYKDHDG DYKDHDIDYK DDDDKGSGSL EVLFQGPMDP IINGNSANVY LTDSYLKGVI SFSECNALGS YIFNGPYLKN DYTNLISRQ NPLIEHMNLK KLNITQSLIS KYHKGEIKLE EPTYFQSLLM TYKSMTSSEQ IATTNLLKKI IRRAIEISDV K VYAILNKL ...String: MGSDYKDHDG DYKDHDIDYK DDDDKGSGSL EVLFQGPMDP IINGNSANVY LTDSYLKGVI SFSECNALGS YIFNGPYLKN DYTNLISRQ NPLIEHMNLK KLNITQSLIS KYHKGEIKLE EPTYFQSLLM TYKSMTSSEQ IATTNLLKKI IRRAIEISDV K VYAILNKL GLKEKDKIKS NNGQDEDNSV ITTIIKDDIL SAVKDNQSHL KADKNHSTKQ KDTIKTTLLK KLMCSMQHPP SW LIHWFNL YTKLNNILTQ YRSNEVKNHG FTLIDNQTLS GFQFILNQYG CIVYHKELKR ITVTTYNQFL TWKDISLSRL NVC LITWIS NCLNTLNKSL GLRCGFNNVI LTQLFLYGDC ILKLFHNEGF YIIKEVEGFI MSLILNITEE DQFRKRFYNS MLNN ITDAA NKAQKNLLSR VCHTLLDKTV SDNIINGRWI ILLSKFLKLI KLAGDNNLNN LSELYFLFRI FGHPMVDERQ AMDAV KINC NETKFYLLSS LSMLRGAFIY RIIKGFVNNY NRWPTLRNAI VLPLRWLTYY KLNTYPSLLE LTERDLIVLS GLRFYR EFR LPKKVDLEMI INDKAISPPK NLIWTSFPRN YMPSHIQNYI EHEKLKFSES DKSRRVLEYY LRDNKFNECD LYNCVVN QS YLNNPNHVVS LTGKERELSV GRMFAMQPGM FRQVQILAEK MIAENILQFF PESLTRYGDL ELQKILELKA GISNKSNR Y NDNYNNYISK CSIITDLSKF NQAFRYETSC ICSDVLDELH GVQSLFSWLH LTIPHVTIIC TYRHAPPYIG DHIVDLNNV DEQSGLYRYH MGGIEGWCQK LWTIEAISLL DLISLKGKFS ITALINGDNQ SIDISKPIRL MEGQTHAQAD YLLALNSLKL LYKEYAGIG HKLKGTETYI SRDMQFMSKT IQHNGVYYPA SIKKVLRVGP WINTILDDFK VSLESIGSLT QELEYRGESL L CSLIFRNV WLYNQIALQL KNHALCNNKL YLDILKVLKH LKTFFNLDNI DTALTLYMNL PMLFGGGDPN LLYRSFYRRT PD FLTEAIV HSVFILSYYT NHDLKDKLQD LSDDRLNKFL TCIITFDKNP NAEFVTLMRD PQALGSERQA KITSEINRLA VTE VLSTAP NKIFSKSAQH YTTTEIDLND IMQNIEPTYP HGLRVVYESL PFYKAEKIVN LISGTKSITN ILEKTSAIDL TDID RATEM MRKNITLLIR ILPLDCNRDK REILSMENLS ITELSKYVRE RSWSLSNIVG VTSPSIMYTM DIKYTTSTIS SGIII EKYN VNSLTRGERG PTKPWVGSST QEKKTMPVYN RQVLTKKQRD QIDLLAKLDW VYASIDNKDE FMEELSIGTL GLTYEK AKK LFPQYLSVNY LHRLTVSSRP CEFPASIPAY RTTNYHFDTS PINRILTEKY GDEDIDIVFQ NCISFGLSLM SVVEQFT NV CPNRIILIPK LNEIHLMKPP IFTGDVDIHK LKQVIQKQHM FLPDKISLTQ YVELF UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human respiratory syncytial virus A2 Human respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 29.062895 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEKFAPEFHG EDANNRATKF LESIKGKFTS PKDPKKKDSI ISVNSIDIEV TKESPITSNS TIINPTNETD DTAGNKPNYQ RKPLVSFKE DPTPSDNPFS KLYKETIETF DNNEEESSYS YEEINDQTND NITARLDRID EKLSEILGML HTLVVASAGP T SARDGIRD ...String: MEKFAPEFHG EDANNRATKF LESIKGKFTS PKDPKKKDSI ISVNSIDIEV TKESPITSNS TIINPTNETD DTAGNKPNYQ RKPLVSFKE DPTPSDNPFS KLYKETIETF DNNEEESSYS YEEINDQTND NITARLDRID EKLSEILGML HTLVVASAGP T SARDGIRD AMIGLREEMI EKIRTEALMT NDRLEAMARL RNEESEKMAK DTSDEVSLNP TSEKLNNLLE GNDSDNDLSL ED FKGENKY FQGHHHHHH UniProtKB: Phosphoprotein |
-Macromolecule #3: 4-(2-aminopropan-2-yl)-N'-[4-(cyclopropyloxy)-3-methoxybenzoyl]-6...
| Macromolecule | Name: 4-(2-aminopropan-2-yl)-N'-[4-(cyclopropyloxy)-3-methoxybenzoyl]-6-(4-fluorophenyl)pyridine-2-carbohydrazide type: ligand / ID: 3 / Number of copies: 1 / Formula: Y6L |
|---|---|
| Molecular weight | Theoretical: 478.515 Da |
| Chemical component information | 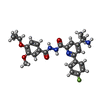 ChemComp-Y6L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 34.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8fpi: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)