[English] 日本語
 Yorodumi
Yorodumi- EMDB-29366: Co-structure of the Human Metapneunomovirus RNA-dependent RNA pol... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Co-structure of the Human Metapneunomovirus RNA-dependent RNA polymerase with MRK-1 | |||||||||
 Map data Map data | Sharpened map used for model refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA-BINDING PROTEIN / HMPV / RDRP / RNA-DEPENDENT RNA POLYMERASE / PRNTASE / POLYRIBONUCLEOTIDYL TRANSFERASE / RNA CAPPING / VIRAL REPLICATION / VIRAL PROTEIN / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / GTPase activity / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Human metapneumovirus CAN97-83 / Human metapneumovirus CAN97-83 /  Human metapneumovirus Human metapneumovirus | |||||||||
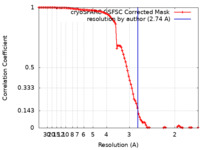
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Fischmann TO | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Conserved allosteric inhibitory site on the respiratory syncytial virus and human metapneumovirus RNA-dependent RNA polymerases. Authors: Victoria A Kleiner / Thierry O Fischmann / John A Howe / Douglas C Beshore / Michael J Eddins / Yan Hou / Todd Mayhood / Daniel Klein / Debbie D Nahas / Bob J Lucas / He Xi / Edward Murray / ...Authors: Victoria A Kleiner / Thierry O Fischmann / John A Howe / Douglas C Beshore / Michael J Eddins / Yan Hou / Todd Mayhood / Daniel Klein / Debbie D Nahas / Bob J Lucas / He Xi / Edward Murray / Daphne Y Ma / Krista Getty / Rachel Fearns /  Abstract: Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) are related RNA viruses responsible for severe respiratory infections and resulting disease in infants, elderly, and ...Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) are related RNA viruses responsible for severe respiratory infections and resulting disease in infants, elderly, and immunocompromised adults. Therapeutic small molecule inhibitors that bind to the RSV polymerase and inhibit viral replication are being developed, but their binding sites and molecular mechanisms of action remain largely unknown. Here we report a conserved allosteric inhibitory site identified on the L polymerase proteins of RSV and HMPV that can be targeted by a dual-specificity, non-nucleoside inhibitor, termed MRK-1. Cryo-EM structures of the inhibitor in complexes with truncated RSV and full-length HMPV polymerase proteins provide a structural understanding of how MRK-1 is active against both viruses. Functional analyses indicate that MRK-1 inhibits conformational changes necessary for the polymerase to engage in RNA synthesis initiation and to transition into an elongation mode. Competition studies reveal that the MRK-1 binding pocket is distinct from that of a capping inhibitor with an overlapping resistance profile, suggesting that the polymerase conformation bound by MRK-1 may be distinct from that involved in mRNA capping. These findings should facilitate optimization of dual RSV and HMPV replication inhibitors and provide insights into the molecular mechanisms underlying their polymerase activities. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29366.map.gz emd_29366.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29366-v30.xml emd-29366-v30.xml emd-29366.xml emd-29366.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29366_fsc.xml emd_29366_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29366.png emd_29366.png | 150.5 KB | ||
| Filedesc metadata |  emd-29366.cif.gz emd-29366.cif.gz | 7.8 KB | ||
| Others |  emd_29366_half_map_1.map.gz emd_29366_half_map_1.map.gz emd_29366_half_map_2.map.gz emd_29366_half_map_2.map.gz | 95.2 MB 95.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29366 http://ftp.pdbj.org/pub/emdb/structures/EMD-29366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29366 | HTTPS FTP |
-Validation report
| Summary document |  emd_29366_validation.pdf.gz emd_29366_validation.pdf.gz | 917.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29366_full_validation.pdf.gz emd_29366_full_validation.pdf.gz | 916.8 KB | Display | |
| Data in XML |  emd_29366_validation.xml.gz emd_29366_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_29366_validation.cif.gz emd_29366_validation.cif.gz | 23.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29366 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29366 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29366 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29366 | HTTPS FTP |
-Related structure data
| Related structure data |  8fpjMC  8fpiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29366.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29366.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map used for model refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Unsharpened half-map
| File | emd_29366_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened half-map
| File | emd_29366_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HUMAN METAPNEUMOVIRUS POLYMERASE (L) PROTEIN BOUND BY THE TETRAME...
| Entire | Name: HUMAN METAPNEUMOVIRUS POLYMERASE (L) PROTEIN BOUND BY THE TETRAMERIC PHOSPHOPROTEIN (P) AND COMPLEXED WITH MRK-1 |
|---|---|
| Components |
|
-Supramolecule #1: HUMAN METAPNEUMOVIRUS POLYMERASE (L) PROTEIN BOUND BY THE TETRAME...
| Supramolecule | Name: HUMAN METAPNEUMOVIRUS POLYMERASE (L) PROTEIN BOUND BY THE TETRAMERIC PHOSPHOPROTEIN (P) AND COMPLEXED WITH MRK-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus CAN97-83 Human metapneumovirus CAN97-83 |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Molecular weight | Theoretical: 235.226891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSDYKDHDG DYKDHDIDYK DDDDKGSGSL EVLFQGPMDP LNESTVNVYL PDSYLKGVIS FSETNAIGSC LLKRPYLKND NTAKVAIEN PVIEHVRLKN AVNSKMKISD YKVVEPVNMQ HEIMKNVHSC ELTLLKQFLT RSKNISTLKL NMICDWLQLK S TSDDTSIL ...String: MGSDYKDHDG DYKDHDIDYK DDDDKGSGSL EVLFQGPMDP LNESTVNVYL PDSYLKGVIS FSETNAIGSC LLKRPYLKND NTAKVAIEN PVIEHVRLKN AVNSKMKISD YKVVEPVNMQ HEIMKNVHSC ELTLLKQFLT RSKNISTLKL NMICDWLQLK S TSDDTSIL SFIDVEFIPS WVSNWFSNWY NLNKLILEFR REEVIRTGSI LCRSLGKLVF IVSSYGCIVK SNKSKRVSFF TY NQLLTWK DVMLSRFNAN FCIWVSNSLN ENQEGLGLRS NLQGMLTNKL YETVDYMLSL CCNEGFSLVK EFEGFIMSEI LRI TEHAQF STRFRNTLLN GLTDQLTKLK NKNRLRVHST VLENNDYPMY EVVLKLLGDT LRCIKLLINK NLENAAELYY IFRI FGHPM VDERDAMDAV KLNNEITKIL RLESLTELRG AFILRIIKGF VDNNKRWPKI KNLKVLSKRW TMYFKAKNYP SQLEL SEQD FLELAAIQFE QEFSVPEKTN LEMVLNDKAI SPPKRLIWSV YPKNYLPETI KNRYLEETFN ASDSLKTRRV LEYYLK DNK FDQKELKSYV VRQEYLNDKE HIVSLTGKER ELSVGRMFAM QPGKQRQIQI LAEKLLADNI VPFFPETLTK YGDLDLQ RI MEIKSELSSI KTRRNDSYNN YIARASIVTD LSKFNQAFRY ETTAICADVA DELHGTQSLF CWLHLIVPMT TMICAYRH A PPETKGEYDI DKIEEQSGLY RYHMGGIEGW CQKLWTMEAI SLLDVVSVKT RCQMTSLLNG DNQSIDVSKP VKLSEGLDE VKADYRLAVK MLKEIRDAYR NIGHKLKEGE TYISRDLQFI SKVIQSEGVM HPTPIKKVLR VGPWINTILD DIKTSAESIG SLCQELEFR GESIIVSLIL RNFWLYNLYM HESKQHPLAG KQLFKQLNKT LTSVQRFFEI KRENEVVDLW MNIPMQFGGG D PVVFYRSF YRRTPDFLTE AISHVDILLK ISANIKNETK VSFFKALLSI EKNERATLTT LMRDPQAVGS ERQAKVTSDI NR TAVTSIL SLSPNQLFSD SAIHYSRNEE EVGIIAENIT PVYPHGLRVL YESLPFHKAE KVVNMISGTK SITNLLQRTS AIN GEDIDR AVSMMLENLG LLSRILSVVV DSIEIPIKSN GRLICCQISR TLRETSWNNM EIVGVTSPSI TTCMDVIYAT SSHL KGIII EKFSTDRTTR GQRGPKSPWV GSSTQEKKLV PVYNRQILSK QQREQLEAIG KMRWVYKGTP GLRRLLNKIC LGSLG ISYK CVKPLLPRFM SVNFLHRLSV SSRPMEFPAS VPAYRTTNYH FDTSPINQAL SERFGNEDIN LVFQNAISCG ISIMSV VEQ LTGRSPKQLV LIPQLEEIDI MPPPVFQGKF NYKLVDKITS DQHIFSPDKI DMLTLGKMLM PTIKGQKTDQ FLNKREN YF HGNNLIESLS AALACHWCGI LTEQCIENNI FKKDWGDGFI SDHAFMDFKI FLCVFKTKLL CSWGSQGKNI KDEDIVDE S IDKLLRIDNT FWRMFSKVMF EPKVKKRIML YDVKFLSLVG YIGFKNWFIE QLRSAELHEI PWIVNAEGDL VEIKSIKIY LQLIEQSLFL RITVLNYTDM AHALTRLIRK KLMCDNALLT PISSPMVNLT QVIDPTTQLD YFPKITFERL KNYDTSSNYA KGKLTRNYM ILLPWQHVNR YNFVFSSTGC KVSLKTCIGK LMKDLNPKVL YFIGEGAGNW MARTACEYPD IKFVYRSLKD D LDHHYPLE YQRVIGELSR IIDSGEGLSM ETTDATQKTH WDLIHRVSKD ALLITLCDAE FKDRDDFFKM VILWRKHVLS CR ICTTYGT DLYLFAKYHA KDCNVKLPFF VRSVATFIMQ GSKLSGSECY ILLTLGHHNS LPCHGEIQNS KMKIAVCNDF YAA KKLDNK SIEANCKSLL SGLRIPINKK ELDRQRRLLT LQSNHSSVAT VGGSKIIESK WLTNKASTII DWLEHILNSP KGEL NYDFF EALENTYPNM IKLIDNLGNA EIKKLIKVTG YMLVSKK UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: Phosphoprotein
| Macromolecule | Name: Phosphoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Molecular weight | Theoretical: 34.814141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFPEGKDIL FMGNEAAKLA EAFQKSLRKP SHKRSQSIIG EKVNTVSETL ELPTISRPTK PTILSEPKLA WTDKGGAIKT EAKQTIKVM DPIEEEEFTE KRVLPSSDGK TPAEKKLKPS TNTKKKVSFT PNEPGKYTKL EKDALDLLSD NEEEDAESSI L TFEERDTS ...String: MSFPEGKDIL FMGNEAAKLA EAFQKSLRKP SHKRSQSIIG EKVNTVSETL ELPTISRPTK PTILSEPKLA WTDKGGAIKT EAKQTIKVM DPIEEEEFTE KRVLPSSDGK TPAEKKLKPS TNTKKKVSFT PNEPGKYTKL EKDALDLLSD NEEEDAESSI L TFEERDTS SLSIEARLES IEEKLSMILG LLRTLNIATA GPTAARDGIR DAMIGIREEL IADIIKEAKG KAAEMMEEEM NQ RTKIGNG SVKLTEKAKE LNKIVEDEST SGESEEEEEL KDTQENNQED DIYQLIMKGE NKYFQGHHHH HHH UniProtKB: Phosphoprotein |
-Macromolecule #3: 4-(2-aminopropan-2-yl)-N'-[4-(cyclopropyloxy)-3-methoxybenzoyl]-6...
| Macromolecule | Name: 4-(2-aminopropan-2-yl)-N'-[4-(cyclopropyloxy)-3-methoxybenzoyl]-6-(4-fluorophenyl)pyridine-2-carbohydrazide type: ligand / ID: 3 / Number of copies: 1 / Formula: Y6L |
|---|---|
| Molecular weight | Theoretical: 478.515 Da |
| Chemical component information | 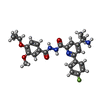 ChemComp-Y6L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8fpj: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)