+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNAPKcs-Ku-DNA subcomplex in PAXX-LR-ATP complex | |||||||||
 Map data Map data | DKB in PAXX-LR-ATP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Kinase / NHEJ / DNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
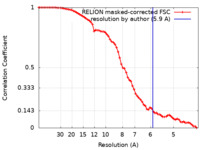
| Method | single particle reconstruction / cryo EM / Resolution: 5.9 Å | |||||||||
 Authors Authors | Chen S / He Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ. Authors: Siyu Chen / Alex Vogt / Linda Lee / Tasmin Naila / Ryan McKeown / Alan E Tomkinson / Susan P Lees-Miller / Yuan He /   Abstract: DNA double-strand breaks (DSBs), one of the most cytotoxic forms of DNA damage, can be repaired by the tightly regulated nonhomologous end joining (NHEJ) machinery (Stinson and Loparo and Zhao ). ...DNA double-strand breaks (DSBs), one of the most cytotoxic forms of DNA damage, can be repaired by the tightly regulated nonhomologous end joining (NHEJ) machinery (Stinson and Loparo and Zhao ). Core NHEJ factors form an initial long-range (LR) synaptic complex that transitions into a DNA-PKcs (DNA-dependent protein kinase, catalytic subunit)-free, short-range state to align the DSB ends (Chen ). Using single-particle cryo-electron microscopy, we have visualized three additional key NHEJ complexes representing different transition states, with DNA-PKcs adopting distinct dimeric conformations within each of them. Upon DNA-PKcs autophosphorylation, the LR complex undergoes a substantial conformational change, with both Ku and DNA-PKcs rotating outward to promote DNA break exposure and DNA-PKcs dissociation. We also captured a dimeric state of catalytically inactive DNA-PKcs, which resembles structures of other PIKK (Phosphatidylinositol 3-kinase-related kinase) family kinases, revealing a model of the full regulatory cycle of DNA-PKcs during NHEJ. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28738.map.gz emd_28738.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28738-v30.xml emd-28738-v30.xml emd-28738.xml emd-28738.xml | 25.5 KB 25.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28738_fsc.xml emd_28738_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28738.png emd_28738.png | 46.9 KB | ||
| Others |  emd_28738_half_map_1.map.gz emd_28738_half_map_1.map.gz emd_28738_half_map_2.map.gz emd_28738_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28738 http://ftp.pdbj.org/pub/emdb/structures/EMD-28738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28738 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28738 | HTTPS FTP |
-Validation report
| Summary document |  emd_28738_validation.pdf.gz emd_28738_validation.pdf.gz | 932 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28738_full_validation.pdf.gz emd_28738_full_validation.pdf.gz | 931.6 KB | Display | |
| Data in XML |  emd_28738_validation.xml.gz emd_28738_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_28738_validation.cif.gz emd_28738_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28738 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28738 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28738 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28738.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28738.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DKB in PAXX-LR-ATP | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.112 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1
| File | emd_28738_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half2
| File | emd_28738_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NHEJ Long-range complex with ATP
| Entire | Name: NHEJ Long-range complex with ATP |
|---|---|
| Components |
|
-Supramolecule #1: NHEJ Long-range complex with ATP
| Supramolecule | Name: NHEJ Long-range complex with ATP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 1.75 MDa |
-Macromolecule #1: XRCC6_HUMAN
| Macromolecule | Name: XRCC6_HUMAN / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MSGWESYYKT EGDEEAEEEQ EENLEASGDY KYSGRDSLIF LVDASKAMFE SQSEDELTPF DMSIQCIQSV YISKIISSDR DLLAVVFYGT EKDKNSVNFK NIYVLQELDN PGAKRILELD QFKGQQGQKR FQDMMGHGSD YSLSEVLWVC ANLFSDVQFK MSHKRIMLFT ...String: MSGWESYYKT EGDEEAEEEQ EENLEASGDY KYSGRDSLIF LVDASKAMFE SQSEDELTPF DMSIQCIQSV YISKIISSDR DLLAVVFYGT EKDKNSVNFK NIYVLQELDN PGAKRILELD QFKGQQGQKR FQDMMGHGSD YSLSEVLWVC ANLFSDVQFK MSHKRIMLFT NEDNPHGNDS AKASRARTKA GDLRDTGIFL DLMHLKKPGG FDISLFYRDI ISIAEDEDLR VHFEESSKLE DLLRKVRAKE TRKRALSRLK LKLNKDIVIS VGIYNLVQKA LKPPPIKLYR ETNEPVKTKT RTFNTSTGGL LLPSDTKRSQ IYGSRQIILE KEETEELKRF DDPGLMLMGF KPLVLLKKHH YLRPSLFVYP EESLVIGSST LFSALLIKCL EKEVAALCRY TPRRNIPPYF VALVPQEEEL DDQKIQVTPP GFQLVFLPFA DDKRKMPFTE KIMATPEQVG KMKAIVEKLR FTYRSDSFEN PVLQQHFRNL EALALDLMEP EQAVDLTLPK VEAMNKRLGS LVDEFKELVY PPDYNPEGKV TKRKHDNEGS GSKRPKVEYS EEELKTHISK GTLGKFTVPM LKEACRAYGL KSGLKKQELL EALTKHFQD |
-Macromolecule #2: XRCC5_HUMAN X-ray repair cross-complementing protein 5
| Macromolecule | Name: XRCC5_HUMAN X-ray repair cross-complementing protein 5 type: protein_or_peptide / ID: 2 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG TDGTDNPLSG GDQYQNITVH RHLMLPDFDL LEDIESKIQP GSQQADFLDA LIVSMDVIQH ETIGKKFEKR HIEIFTDLSS RFSKSQLDII IHSLKKCDIS LQFFLPFSLG ...String: MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG TDGTDNPLSG GDQYQNITVH RHLMLPDFDL LEDIESKIQP GSQQADFLDA LIVSMDVIQH ETIGKKFEKR HIEIFTDLSS RFSKSQLDII IHSLKKCDIS LQFFLPFSLG KEDGSGDRGD GPFRLGGHGP SFPLKGITEQ QKEGLEIVKM VMISLEGEDG LDEIYSFSES LRKLCVFKKI ERHSIHWPCR LTIGSNLSIR IAAYKSILQE RVKKTWTVVD AKTLKKEDIQ KETVYCLNDD DETEVLKEDI IQGFRYGSDI VPFSKVDEEQ MKYKSEGKCF SVLGFCKSSQ VQRRFFMGNQ VLKVFAARDD EAAAVALSSL IHALDDLDMV AIVRYAYDKR ANPQVGVAFP HIKHNYECLV YVQLPFMEDL RQYMFSSLKN SKKYAPTEAQ LNAVDALIDS MSLAKKDEKT DTLEDLFPTT KIPNPRFQRL FQCLLHRALH PREPLPPIQQ HIWNMLNPPA EVTTKSQIPL SKIKTLFPLI EAKKKDQVTA QEIFQDNHED GPTAKKLKTE QGGAHFSVSS LAEGSVTSVG SVNPAENFRV LVKQKKASFE EASNQLINHI EQFLDTNETP YFMKSIDCIR AFREEAIKFS EEQRFNNFLK ALQEKVEIKQ LNHFWEIVVQ DGITLITKEE ASGSSVTAEE AKKFLAPKDK PSGDTAAVFE EGGDVDDLLD MI |
-Macromolecule #3: PRKDC_HUMAN DNA-dependent protein kinase catalytic subunit
| Macromolecule | Name: PRKDC_HUMAN DNA-dependent protein kinase catalytic subunit type: protein_or_peptide / ID: 3 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAGSGAGVRC SLLRLQETLS AADRCGAALA GHQLIRGLGQ ECVLSSSPAV LALQTSLVFS RDFGLLVFVR KSLNSIEFRE CREEILKFLC IFLEKMGQKI APYSVEIKNT CTSVYTKDRA AKCKIPALDL LIKLLQTFRS SRLMDEFKIG ELFSKFYGEL ALKKKIPDTV ...String: MAGSGAGVRC SLLRLQETLS AADRCGAALA GHQLIRGLGQ ECVLSSSPAV LALQTSLVFS RDFGLLVFVR KSLNSIEFRE CREEILKFLC IFLEKMGQKI APYSVEIKNT CTSVYTKDRA AKCKIPALDL LIKLLQTFRS SRLMDEFKIG ELFSKFYGEL ALKKKIPDTV LEKVYELLGL LGEVHPSEMI NNAENLFRAF LGELKTQMTS AVREPKLPVL AGCLKGLSSL LCNFTKSMEE DPQTSREIFN FVLKAIRPQI DLKRYAVPSA GLRLFALHAS QFSTCLLDNY VSLFEVLLKW CAHTNVELKK AALSALESFL KQVSNMVAKN AEMHKNKLQY FMEQFYGIIR NVDSNNKELS IAIRGYGLFA GPCKVINAKD VDFMYVELIQ RCKQMFLTQT DTGDDRVYQM PSFLQSVASV LLYLDTVPEV YTPVLEHLVV MQIDSFPQYS PKMQLVCCRA IVKVFLALAA KGPVLRNCIS TVVHQGLIRI CSKPVVLPKG PESESEDHRA SGEVRTGKWK VPTYKDYVDL FRHLLSSDQM MDSILADEAF FSVNSSSESL NHLLYDEFVK SVLKIVEKLD LTLEIQTVGE QENGDEAPGV WMIPTSDPAA NLHPAKPKDF SAFINLVEFC REILPEKQAE FFEPWVYSFS YELILQSTRL PLISGFYKLL SITVRNAKKI KYFEGVSPKS LKHSPEDPEK YSCFALFVKF GKEVAVKMKQ YKDELLASCL TFLLSLPHNI IELDVRAYVP ALQMAFKLGL SYTPLAEVGL NALEEWSIYI DRHVMQPYYK DILPCLDGYL KTSALSDETK NNWEVSALSR AAQKGFNKVV LKHLKKTKNL SSNEAISLEE IRIRVVQMLG SLGGQINKNL LTVTSSDEMM KSYVAWDREK RLSFAVPFRE MKPVIFLDVF LPRVTELALT ASDRQTKVAA CELLHSMVMF MLGKATQMPE GGQGAPPMYQ LYKRTFPVLL RLACDVDQVT RQLYEPLVMQ LIHWFTNNKK FESQDTVALL EAILDGIVDP VDSTLRDFCG RCIREFLKWS IKQITPQQQE KSPVNTKSLF KRLYSLALHP NAFKRLGASL AFNNIYREFR EEESLVEQFV FEALVIYMES LALAHADEKS LGTIQQCCDA IDHLCRIIEK KHVSLNKAKK RRLPRGFPPS ASLCLLDLVK WLLAHCGRPQ TECRHKSIEL FYKFVPLLPG NRSPNLWLKD VLKEEGVSFL INTFEGGGCG QPSGILAQPT LLYLRGPFSL QATLCWLDLL LAALECYNTF IGERTVGALQ VLGTEAQSSL LKAVAFFLES IAMHDIIAAE KCFGTGAAGN RTSPQEGERY NYSKCTVVVR IMEFTTTLLN TSPEGWKLLK KDLCNTHLMR VLVQTLCEPA SIGFNIGDVQ VMAHLPDVCV NLMKALKMSP YKDILETHLR EKITAQSIEE LCAVNLYGPD AQVDRSRLAA VVSACKQLHR AGLLHNILPS QSTDLHHSVG TELLSLVYKG IAPGDERQCL PSLDLSCKQL ASGLLELAFA FGGLCERLVS LLLNPAVLST ASLGSSQGSV IHFSHGEYFY SLFSETINTE LLKNLDLAVL ELMQSSVDNT KMVSAVLNGM LDQSFRERAN QKHQGLKLAT TILQHWKKCD SWWAKDSPLE TKMAVLALLA KILQIDSSVS FNTSHGSFPE VFTTYISLLA DTKLDLHLKG QAVTLLPFFT SLTGGSLEEL RRVLEQLIVA HFPMQSREFP PGTPRFNNYV DCMKKFLDAL ELSQSPMLLE LMTEVLCREQ QHVMEELFQS SFRRIARRGS CVTQVGLLES VYEMFRKDDP RLSFTRQSFV DRSLLTLLWH CSLDALREFF STIVVDAIDV LKSRFTKLNE STFDTQITKK MGYYKILDVM YSRLPKDDVH AKESKINQVF HGSCITEGNE LTKTLIKLCY DAFTENMAGE NQLLERRRLY HCAAYNCAIS VICCVFNELK FYQGFLFSEK PEKNLLIFEN LIDLKRRYNF PVEVEVPMER KKKYIEIRKE AREAANGDSD GPSYMSSLSY LADSTLSEEM SQFDFSTGVQ SYSYSSQDPR PATGRFRRRE QRDPTVHDDV LELEMDELNR HECMAPLTAL VKHMHRSLGP PQGEEDSVPR DLPSWMKFLH GKLGNPIVPL NIRLFLAKLV INTEEVFRPY AKHWLSPLLQ LAASENNGGE GIHYMVVEIV ATILSWTGLA TPTGVPKDEV LANRLLNFLM KHVFHPKRAV FRHNLEIIKT LVECWKDCLS IPYRLIFEKF SGKDPNSKDN SVGIQLLGIV MANDLPPYDP QCGIQSSEYF QALVNNMSFV RYKEVYAAAA EVLGLILRYV MERKNILEES LCELVAKQLK QHQNTMEDKF IVCLNKVTKS FPPLADRFMN AVFFLLPKFH GVLKTLCLEV VLCRVEGMTE LYFQLKSKDF VQVMRHRDDE RQKVCLDIIY KMMPKLKPVE LRELLNPVVE FVSHPSTTCR EQMYNILMWI HDNYRDPESE TDNDSQEIFK LAKDVLIQGL IDENPGLQLI IRNFWSHETR LPSNTLDRLL ALNSLYSPKI EVHFLSLATN FLLEMTSMSP DYPNPMFEHP LSECEFQEYT IDSDWRFRST VLTPMFVETQ ASQGTLQTRT QEGSLSARWP VAGQIRATQQ QHDFTLTQTA DGRSSFDWLT GSSTDPLVDH TSPSSDSLLF AHKRSERLQR APLKSVGPDF GKKRLGLPGD EVDNKVKGAA GRTDLLRLRR RFMRDQEKLS LMYARKGVAE QKREKEIKSE LKMKQDAQVV LYRSYRHGDL PDIQIKHSSL ITPLQAVAQR DPIIAKQLFS SLFSGILKEM DKFKTLSEKN NITQKLLQDF NRFLNTTFSF FPPFVSCIQD ISCQHAALLS LDPAAVSAGC LASLQQPVGI RLLEEALLRL LPAELPAKRV RGKARLPPDV LRWVELAKLY RSIGEYDVLR GIFTSEIGTK QITQSALLAE ARSDYSEAAK QYDEALNKQD WVDGEPTEAE KDFWELASLD CYNHLAEWKS LEYCSTASID SENPPDLNKI WSEPFYQETY LPYMIRSKLK LLLQGEADQS LLTFIDKAMH GELQKAILEL HYSQELSLLY LLQDDVDRAK YYIQNGIQSF MQNYSSIDVL LHQSRLTKLQ SVQALTEIQE FISFISKQGN LSSQVPLKRL LNTWTNRYPD AKMDPMNIWD DIITNRCFFL SKIEEKLTPL PEDNSMNVDQ DGDPSDRMEV QEQEEDISSL IRSCKFSMKM KMIDSARKQN NFSLAMKLLK ELHKESKTRD DWLVSWVQSY CRLSHCRSRS QGCSEQVLTV LKTVSLLDEN NVSSYLSKNI LAFRDQNILL GTTYRIIANA LSSEPACLAE IEEDKARRIL ELSGSSSEDS EKVIAGLYQR AFQHLSEAVQ AAEEEAQPPS WSCGPAAGVI DAYMTLADFC DQQLRKEEEN ASVIDSAELQ AYPALVVEKM LKALKLNSNE ARLKFPRLLQ IIERYPEETL SLMTKEISSV PCWQFISWIS HMVALLDKDQ AVAVQHSVEE ITDNYPQAIV YPFIISSESY SFKDTSTGHK NKEFVARIKS KLDQGGVIQD FINALDQLSN PELLFKDWSN DVRAELAKTP VNKKNIEKMY ERMYAALGDP KAPGLGAFRR KFIQTFGKEF DKHFGKGGSK LLRMKLSDFN DITNMLLLKM NKDSKPPGNL KECSPWMSDF KVEFLRNELE IPGQYDGRGK PLPEYHVRIA GFDERVTVMA SLRRPKRIII RGHDEREHPF LVKGGEDLRQ DQRVEQLFQV MNGILAQDSA CSQRALQLRT YSVVPMTSRL GLIEWLENTV TLKDLLLNTM SQEEKAAYLS DPRAPPCEYK DWLTKMSGKH DVGAYMLMYK GANRTETVTS FRKRESKVPA DLLKRAFVRM STSPEAFLAL RSHFASSHAL ICISHWILGI GDRHLNNFMV AMETGGVIGI DFGHAFGSAT QFLPVPELMP FRLTRQFINL MLPMKETGLM YSIMVHALRA FRSDPGLLTN TMDVFVKEPS FDWKNFEQKM LKKGGSWIQE INVAEKNWYP RQKICYAKRK LAGANPAVIT CDELLLGHEK APAFRDYVAV ARGSKDHNIR AQEPESGLSE ETQVKCLMDQ ATDPNILGRT WEGWEPWM |
-Macromolecule #4: DNA(31-MER)
| Macromolecule | Name: DNA(31-MER) / type: dna / ID: 4 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (DT)(DC)(DT)(DA)(DA)(DG)(DA)(DA)(DC)(DT) (DC)(DT)(DG)(DA)(DT)(DG)(DT)(DC)(DA)(DG) (DT)(DA)(DG)(DA)(DT)(DT)(DA)(DC)(DA)(DC) (DT) |
-Macromolecule #5: DNA(30-MER)
| Macromolecule | Name: DNA(30-MER) / type: dna / ID: 5 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (DG)(DT)(DG)(DT)(DA)(DA)(DT)(DC)(DT)(DA) (DC)(DT)(DG)(DA)(DC)(DA)(DT)(DC)(DA)(DG) (DA)(DG)(DT)(DT)(DC)(DT)(DT)(DA)(DG) (DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 302 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 30448 / Average exposure time: 4.0 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 4.0 µm / Calibrated defocus min: 2.0 µm / Calibrated magnification: 60000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
|---|
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)