+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | XRCC4-XLF-XRCC4 subcomplex of PAXX-LR complex | |||||||||
 Map data Map data | XXL in PAXX-LR | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Kinase / NHEJ / DNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 14.0 Å | |||||||||
 Authors Authors | Chen S / He Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ. Authors: Siyu Chen / Alex Vogt / Linda Lee / Tasmin Naila / Ryan McKeown / Alan E Tomkinson / Susan P Lees-Miller / Yuan He /   Abstract: DNA double-strand breaks (DSBs), one of the most cytotoxic forms of DNA damage, can be repaired by the tightly regulated nonhomologous end joining (NHEJ) machinery (Stinson and Loparo and Zhao ). ...DNA double-strand breaks (DSBs), one of the most cytotoxic forms of DNA damage, can be repaired by the tightly regulated nonhomologous end joining (NHEJ) machinery (Stinson and Loparo and Zhao ). Core NHEJ factors form an initial long-range (LR) synaptic complex that transitions into a DNA-PKcs (DNA-dependent protein kinase, catalytic subunit)-free, short-range state to align the DSB ends (Chen ). Using single-particle cryo-electron microscopy, we have visualized three additional key NHEJ complexes representing different transition states, with DNA-PKcs adopting distinct dimeric conformations within each of them. Upon DNA-PKcs autophosphorylation, the LR complex undergoes a substantial conformational change, with both Ku and DNA-PKcs rotating outward to promote DNA break exposure and DNA-PKcs dissociation. We also captured a dimeric state of catalytically inactive DNA-PKcs, which resembles structures of other PIKK (Phosphatidylinositol 3-kinase-related kinase) family kinases, revealing a model of the full regulatory cycle of DNA-PKcs during NHEJ. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28736.map.gz emd_28736.map.gz | 39.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28736-v30.xml emd-28736-v30.xml emd-28736.xml emd-28736.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28736.png emd_28736.png | 19.2 KB | ||
| Others |  emd_28736_half_map_1.map.gz emd_28736_half_map_1.map.gz emd_28736_half_map_2.map.gz emd_28736_half_map_2.map.gz | 32.5 MB 32.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28736 http://ftp.pdbj.org/pub/emdb/structures/EMD-28736 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28736 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28736 | HTTPS FTP |
-Validation report
| Summary document |  emd_28736_validation.pdf.gz emd_28736_validation.pdf.gz | 754.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28736_full_validation.pdf.gz emd_28736_full_validation.pdf.gz | 753.7 KB | Display | |
| Data in XML |  emd_28736_validation.xml.gz emd_28736_validation.xml.gz | 11 KB | Display | |
| Data in CIF |  emd_28736_validation.cif.gz emd_28736_validation.cif.gz | 12.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28736 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28736 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28736 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28736 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28736.map.gz / Format: CCP4 / Size: 43.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28736.map.gz / Format: CCP4 / Size: 43.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | XXL in PAXX-LR | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.158 Å | ||||||||||||||||||||||||||||||||||||
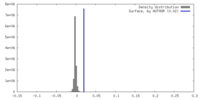
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half1
| File | emd_28736_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
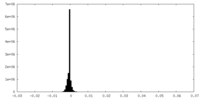
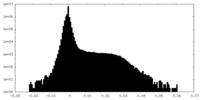
| Density Histograms |
-Half map: half2
| File | emd_28736_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
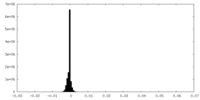
| Density Histograms |
- Sample components
Sample components
-Entire : NHEJ Long-range complex with PAXX
| Entire | Name: NHEJ Long-range complex with PAXX |
|---|---|
| Components |
|
-Supramolecule #1: NHEJ Long-range complex with PAXX
| Supramolecule | Name: NHEJ Long-range complex with PAXX / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.75 MDa |
-Macromolecule #1: XRCC4_HUMAN DNA repair protein XRCC4
| Macromolecule | Name: XRCC4_HUMAN DNA repair protein XRCC4 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MERKISRIHL VSEPSITHFL QVSWEKTLES GFVITLTDGH SAWTGTVSES EISQEADDMA MEKGKYVGEL RKALLSGAGP ADVYTFNFSK ESCYFFFEKN LKDVSFRLGS FNLEKVENPA EVIRELICYC LDTIAENQAK NEHLQKENER LLRDWNDVQG RFEKCVSAKE ...String: MERKISRIHL VSEPSITHFL QVSWEKTLES GFVITLTDGH SAWTGTVSES EISQEADDMA MEKGKYVGEL RKALLSGAGP ADVYTFNFSK ESCYFFFEKN LKDVSFRLGS FNLEKVENPA EVIRELICYC LDTIAENQAK NEHLQKENER LLRDWNDVQG RFEKCVSAKE ALETDLYKRF ILVLNEKKTK IRSLHNKLLN AAQEREKDIK QEGETAICSE MTADRDPVYD ESTDEESENQ TDLSGLASAA VSKDDSIISS LDVTDIAPSR KRRQRMQRNL GTEPKMAPQE NQLQEKENSR PDSSLPETSK KEHISAENMS LETLRNSSPE DLFDEI |
-Macromolecule #2: NHEJ1_HUMAN Non-homologous end-joining factor 1
| Macromolecule | Name: NHEJ1_HUMAN Non-homologous end-joining factor 1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MEELEQGLLM QPWAWLQLAE NSLLAKVFIT KQGYALLVSD LQQVWHEQVD TSVVSQRAKE LNKRLTAPPA AFLCHLDNLL RPLLKDAAHP SEATFSCDCV ADALILRVRS ELSGLPFYWN FHCMLASPSL VSQHLIRPLM GMSLALQCQV RELATLLHMK DLEIQDYQES ...String: MEELEQGLLM QPWAWLQLAE NSLLAKVFIT KQGYALLVSD LQQVWHEQVD TSVVSQRAKE LNKRLTAPPA AFLCHLDNLL RPLLKDAAHP SEATFSCDCV ADALILRVRS ELSGLPFYWN FHCMLASPSL VSQHLIRPLM GMSLALQCQV RELATLLHMK DLEIQDYQES GATLIRDRLK TEPFEENSFL EQFMIEKLPE ACSIGDGKPF VMNLQDLYMA VTTQEVQVGQ KHQGAGDPHT SNSASLQGID SQCVNQPEQL VSSAPTLSAP EKESTGTSGP LQRPQLSKVK RKKPRGLFS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 302 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 11006 / Average exposure time: 4.0 sec. / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 4.0 µm / Calibrated defocus min: 2.0 µm / Calibrated magnification: 60000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)