+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2609 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | KIF14 Motor Domain Microtubule complex | |||||||||
 Map data Map data | kinesin KIF14 mtor domain micerotubule complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinesin / Micotubule / Tubulin / KIF14 / Kinesin-3 | |||||||||
| Biological species |   | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 15.5 Å | |||||||||
 Authors Authors | Arora K / Talje L / Asenjo AB / Andersen P / Atchia K / Joshi M / Sosa H / Allingham JS / Kwok BH | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2014 Journal: J Mol Biol / Year: 2014Title: KIF14 binds tightly to microtubules and adopts a rigor-like conformation. Authors: Kritica Arora / Lama Talje / Ana B Asenjo / Parker Andersen / Kaleem Atchia / Monika Joshi / Hernando Sosa / John S Allingham / Benjamin H Kwok /   Abstract: The mitotic kinesin motor protein KIF14 is essential for cytokinesis during cell division and has been implicated in cerebral development and a variety of human cancers. Here we show that the mouse ...The mitotic kinesin motor protein KIF14 is essential for cytokinesis during cell division and has been implicated in cerebral development and a variety of human cancers. Here we show that the mouse KIF14 motor domain binds tightly to microtubules and does not display typical nucleotide-dependent changes in this affinity. It also has robust ATPase activity but very slow motility. A crystal structure of the ADP-bound form of the KIF14 motor domain reveals a dramatically opened ATP-binding pocket, as if ready to exchange its bound ADP for Mg·ATP. In this state, the central β-sheet is twisted ~10° beyond the maximal amount observed in other kinesins. This configuration has only been seen in the nucleotide-free states of myosins-known as the "rigor-like" state. Fitting of this atomic model to electron density maps from cryo-electron microscopy indicates a distinct binding configuration of the motor domain to microtubules. We postulate that these properties of KIF14 are well suited for stabilizing midbody microtubules during cytokinesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2609.map.gz emd_2609.map.gz | 877.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2609-v30.xml emd-2609-v30.xml emd-2609.xml emd-2609.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd-2609-Kif14_mapview.JPG emd-2609-Kif14_mapview.JPG | 189.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2609 http://ftp.pdbj.org/pub/emdb/structures/EMD-2609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2609 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2609 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2609.map.gz / Format: CCP4 / Size: 918.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2609.map.gz / Format: CCP4 / Size: 918.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | kinesin KIF14 mtor domain micerotubule complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
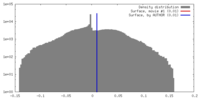
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Kinesin KIF14 motor domain microtubule complex
| Entire | Name: Kinesin KIF14 motor domain microtubule complex |
|---|---|
| Components |
|
-Supramolecule #1000: Kinesin KIF14 motor domain microtubule complex
| Supramolecule | Name: Kinesin KIF14 motor domain microtubule complex / type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Macromolecule #1: Kinesin KIF14 motor domain
| Macromolecule | Name: Kinesin KIF14 motor domain / type: protein_or_peptide / ID: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #2: Tubulin
| Macromolecule | Name: Tubulin / type: protein_or_peptide / ID: 2 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK I |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Date | Nov 25, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number real images: 30 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | Initial reference map obtained by Fourier-Bessel reconstruction as implemented in SUPRIM & PHOELIX Single particle alignment, 3D reconstruction and refinement done using IHRSR, SPIDER and custom routines. |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.40237 Å Applied symmetry - Helical parameters - Δ&Phi: 168.08788 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 15.5 Å / Resolution method: OTHER Software - Name: SUPRIM, PHOELIX, IHRSR, SPIDER, CTFFIND3, CUSTOM, (emglue.py) |
| CTF correction | Details: Each Particle |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The 2 domains (tubulin heterodimer and KIF14 motor domain) were separately fitted. 1 nm resolution density maps of the atomic models were fitted within the cryo-em map using the global fit option of the fitmap command of the UCSF-CHIMERA program. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)