[English] 日本語
 Yorodumi
Yorodumi- EMDB-1975: Functional reconstitution of the 13-subunit human translation ini... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1975 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Functional reconstitution of the 13-subunit human translation initiation factor eIF3. | |||||||||
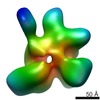
 Map data Map data | eIF3 PCI MPN core negative stain reconstruction | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | translation initiation / eukaryotic initiation factor 3 / ribosome / hepatitis C virus / internal ribosome entry site / proteasome / COP9 signalosome | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Querol-Audi J / Sun C / Todorovic A / Bai Y / Villa N / Snyder M / Ashchyan J / Lewis C / Hartland A / Gradia S ...Querol-Audi J / Sun C / Todorovic A / Bai Y / Villa N / Snyder M / Ashchyan J / Lewis C / Hartland A / Gradia S / Fraser CS / Doudna JA / Nogales E / Cate JHD | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Authors: Chaomin Sun / Aleksandar Todorovic / Jordi Querol-Audí / Yun Bai / Nancy Villa / Monica Snyder / John Ashchyan / Christopher S Lewis / Abbey Hartland / Scott Gradia / Christopher S Fraser / ...Authors: Chaomin Sun / Aleksandar Todorovic / Jordi Querol-Audí / Yun Bai / Nancy Villa / Monica Snyder / John Ashchyan / Christopher S Lewis / Abbey Hartland / Scott Gradia / Christopher S Fraser / Jennifer A Doudna / Eva Nogales / Jamie H D Cate /  Abstract: Protein fate in higher eukaryotes is controlled by three complexes that share conserved architectural elements: the proteasome, COP9 signalosome, and eukaryotic translation initiation factor 3 (eIF3). ...Protein fate in higher eukaryotes is controlled by three complexes that share conserved architectural elements: the proteasome, COP9 signalosome, and eukaryotic translation initiation factor 3 (eIF3). Here we reconstitute the 13-subunit human eIF3 in Escherichia coli, revealing its structural core to be the eight subunits with conserved orthologues in the proteasome lid complex and COP9 signalosome. This structural core in eIF3 binds to the small (40S) ribosomal subunit, to translation initiation factors involved in mRNA cap-dependent initiation, and to the hepatitis C viral (HCV) internal ribosome entry site (IRES) RNA. Addition of the remaining eIF3 subunits enables reconstituted eIF3 to assemble intact initiation complexes with the HCV IRES. Negative-stain EM reconstructions of reconstituted eIF3 further reveal how the approximately 400 kDa molecular mass structural core organizes the highly flexible 800 kDa molecular mass eIF3 complex, and mediates translation initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1975.map.gz emd_1975.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1975-v30.xml emd-1975-v30.xml emd-1975.xml emd-1975.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  8mer_emd1975.jpg 8mer_emd1975.jpg | 20.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1975 http://ftp.pdbj.org/pub/emdb/structures/EMD-1975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1975 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1975.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1975.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | eIF3 PCI MPN core negative stain reconstruction | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PCI MPN core of the human eukaryotic translation initiation facto...
| Entire | Name: PCI MPN core of the human eukaryotic translation initiation factor eIF3 |
|---|---|
| Components |
|
-Supramolecule #1000: PCI MPN core of the human eukaryotic translation initiation facto...
| Supramolecule | Name: PCI MPN core of the human eukaryotic translation initiation factor eIF3 type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: Subcomplex containing 8 core subunits / Number unique components: 8 |
|---|---|
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: eIF3 a
| Macromolecule | Name: eIF3 a / type: protein_or_peptide / ID: 1 / Name.synonym: eIF3 a / Details: truncated version, contains residues 5-654 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Cell: n/atruncated version, contains residues 302-913 Homo sapiens (human) / synonym: Human / Cell: n/atruncated version, contains residues 302-913 |
| Recombinant expression | Organism:  |
-Macromolecule #2: eIF3 c
| Macromolecule | Name: eIF3 c / type: protein_or_peptide / ID: 2 / Name.synonym: eIF3 c / Details: truncated version, contains residues 302-913 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Macromolecule #3: eIF3 e
| Macromolecule | Name: eIF3 e / type: protein_or_peptide / ID: 3 / Name.synonym: eIF3 e / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Macromolecule #4: eIF3 f
| Macromolecule | Name: eIF3 f / type: protein_or_peptide / ID: 4 / Name.synonym: eIF3 f / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Macromolecule #5: eIF3 h
| Macromolecule | Name: eIF3 h / type: protein_or_peptide / ID: 5 / Name.synonym: eIF3 h / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Macromolecule #6: eIF3 k
| Macromolecule | Name: eIF3 k / type: protein_or_peptide / ID: 6 / Name.synonym: eIF3 k / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Macromolecule #7: eIF3 l
| Macromolecule | Name: eIF3 l / type: protein_or_peptide / ID: 7 / Name.synonym: eIF3 l / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Macromolecule #8: eIF3 m
| Macromolecule | Name: eIF3 m / type: protein_or_peptide / ID: 8 / Name.synonym: eIF3 m / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM HEPES, 120 mM KCl, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, 3% trehalose |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein stained with 3% w/v uranyl acetate for 40 seconds |
| Grid | Details: 200 mesh Cu grid |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 100,000 times magnification |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS (4k x 4k) / Number real images: 455 / Average electron dose: 20 e/Å2 / Details: Data collected using TVIPS camera. |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 6.3 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.63 µm / Nominal magnification: 49000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: SIDE ENTRY, EUCENTRIC |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















