[English] 日本語
 Yorodumi
Yorodumi- EMDB-25839: 1.78A reconstruction of mouse heavy chain apoferritin collected a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25839 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1.78A reconstruction of mouse heavy chain apoferritin collected at 500 micrographs/hr | |||||||||
 Map data Map data | main sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | METAL TRANSPORT | |||||||||
| Biological species |  | |||||||||
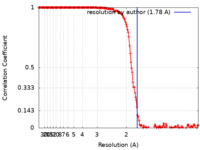
| Method | single particle reconstruction / cryo EM / Resolution: 1.78 Å | |||||||||
 Authors Authors | Peck JV / Fay JF / Strauss JD | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2022 Journal: IUCrJ / Year: 2022Title: High-speed high-resolution data collection on a 200 keV cryo-TEM. Authors: Jared V Peck / Jonathan F Fay / Joshua D Strauss /  Abstract: Limitations to successful single-particle cryo-electron microscopy (cryo-EM) projects include stable sample generation, production of quality cryo-EM grids with randomly oriented particles embedded ...Limitations to successful single-particle cryo-electron microscopy (cryo-EM) projects include stable sample generation, production of quality cryo-EM grids with randomly oriented particles embedded in thin vitreous ice and access to microscope time. To address the limitation of microscope time, methodologies to more efficiently collect data on a 200 keV Talos Arctica cryo-transmission electron microscope at speeds as fast as 720 movies per hour (∼17 000 per day) were tested. In this study, key parameters were explored to increase data collection speed including: (1) using the beam-image shift method to acquire multiple images per stage position, (2) employing UltrAufoil TEM grids with R0.6/1 hole spacing, (3) collecting hardware-binned data and (4) adjusting the image shift delay factor in . Here, eight EM maps of mouse apoferritin at 1.8-1.9 Å resolution were obtained in the analysis with data collection times for each dataset ranging from 56 min to 2 h. An EM map of mouse apoferritin at 1.78 Å was obtained from an overnight data collection at a speed of 500 movies per hour and subgroup analysis performed, with no significant variation observed in data quality by image shift distance and image shift delay. The findings and operating procedures detailed herein allow for rapid turnover of single-particle cryo-EM structure determination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
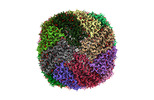
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25839.map.gz emd_25839.map.gz | 398.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25839-v30.xml emd-25839-v30.xml emd-25839.xml emd-25839.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25839_fsc.xml emd_25839_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_25839.png emd_25839.png | 95.8 KB | ||
| Filedesc metadata |  emd-25839.cif.gz emd-25839.cif.gz | 3.6 KB | ||
| Others |  emd_25839_half_map_1.map.gz emd_25839_half_map_1.map.gz emd_25839_half_map_2.map.gz emd_25839_half_map_2.map.gz | 391.2 MB 391.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25839 http://ftp.pdbj.org/pub/emdb/structures/EMD-25839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25839 | HTTPS FTP |
-Validation report
| Summary document |  emd_25839_validation.pdf.gz emd_25839_validation.pdf.gz | 1000.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25839_full_validation.pdf.gz emd_25839_full_validation.pdf.gz | 999.9 KB | Display | |
| Data in XML |  emd_25839_validation.xml.gz emd_25839_validation.xml.gz | 25.2 KB | Display | |
| Data in CIF |  emd_25839_validation.cif.gz emd_25839_validation.cif.gz | 32.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25839 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25839 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25839 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25839.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25839.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main sharp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.545 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half
| File | emd_25839_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half | ||||||||||||
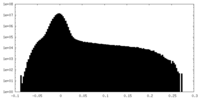
| Projections & Slices |
| ||||||||||||
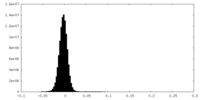
| Density Histograms |
-Half map: other half
| File | emd_25839_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | other half | ||||||||||||
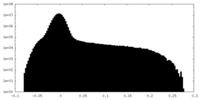
| Projections & Slices |
| ||||||||||||
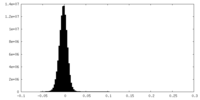
| Density Histograms |
- Sample components
Sample components
-Entire : mouse heavy chain apoferritin
| Entire | Name: mouse heavy chain apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: mouse heavy chain apoferritin
| Supramolecule | Name: mouse heavy chain apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 487.32 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z
Z Y
Y X
X