+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24321 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ADP state actin filament | ||||||||||||
 Map data Map data | Cryo-EM structure of F actin in the ADP state | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationStriated Muscle Contraction / striated muscle thin filament / skeletal muscle thin filament assembly / skeletal muscle fiber development / stress fiber / actin filament / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin cytoskeleton / hydrolase activity / ATP binding Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
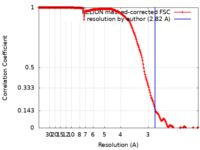
| Method | helical reconstruction / cryo EM / Resolution: 2.82 Å | ||||||||||||
 Authors Authors | Gong R / Espinosa de los Reyes S / Reynolds MJ / Gurel P / Alushin GM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia. Authors: Rui Gong / Fangfang Jiang / Zane G Moreland / Matthew J Reynolds / Santiago Espinosa de Los Reyes / Pinar Gurel / Arik Shams / James B Heidings / Michael R Bowl / Jonathan E Bird / Gregory M Alushin /   Abstract: The motor protein myosin-15 is necessary for the development and maintenance of mechanosensory stereocilia, and mutations in myosin-15 cause hereditary deafness. In addition to transporting actin ...The motor protein myosin-15 is necessary for the development and maintenance of mechanosensory stereocilia, and mutations in myosin-15 cause hereditary deafness. In addition to transporting actin regulatory machinery to stereocilia tips, myosin-15 directly nucleates actin filament ("F-actin") assembly, which is disrupted by a progressive hearing loss mutation (p.D1647G, ""). Here, we present cryo-electron microscopy structures of myosin-15 bound to F-actin, providing a framework for interpreting the impacts of deafness mutations on motor activity and actin nucleation. Rigor myosin-15 evokes conformational changes in F-actin yet maintains flexibility in actin's D-loop, which mediates inter-subunit contacts, while the mutant locks the D-loop in a single conformation. Adenosine diphosphate-bound myosin-15 also locks the D-loop, which correspondingly blunts actin-polymerization stimulation. We propose myosin-15 enhances polymerization by bridging actin protomers, regulating nucleation efficiency by modulating actin's structural plasticity in a myosin nucleotide state-dependent manner. This tunable regulation of actin polymerization could be harnessed to precisely control stereocilium height. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Structural basis for tunable control of actin dynamics by myosin-15 in mechanosensory stereocilia Authors: Gong R / Jiang F / Moreland ZG / Reynolds MJ / Gurel PS / Shams A / Bowl MR / Bird JE / Alushin GM | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24321.map.gz emd_24321.map.gz | 287.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24321-v30.xml emd-24321-v30.xml emd-24321.xml emd-24321.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24321_fsc.xml emd_24321_fsc.xml | 18 KB | Display |  FSC data file FSC data file |
| Images |  emd_24321.png emd_24321.png | 54.9 KB | ||
| Masks |  emd_24321_msk_1.map emd_24321_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_24321_half_map_1.map.gz emd_24321_half_map_1.map.gz emd_24321_half_map_2.map.gz emd_24321_half_map_2.map.gz | 410.7 MB 410.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24321 http://ftp.pdbj.org/pub/emdb/structures/EMD-24321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24321 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24321 | HTTPS FTP |
-Related structure data
| Related structure data |  7r8vMC  7r91C  7rb8C  7rb9C  7udtC  7uduC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24321.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24321.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of F actin in the ADP state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
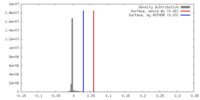
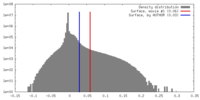
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24321_msk_1.map emd_24321_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: cryo-EM structure of F actin in the ADP state
| File | emd_24321_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM structure of F actin in the ADP state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: cryo-EM structure of F actin in the ADP state
| File | emd_24321_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM structure of F actin in the ADP state | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : filamentous chicken skeletal muscle actin
| Entire | Name: filamentous chicken skeletal muscle actin |
|---|---|
| Components |
|
-Supramolecule #1: filamentous chicken skeletal muscle actin
| Supramolecule | Name: filamentous chicken skeletal muscle actin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: chicken skeletal muscle actin purified from fresh chicken breast |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.4 kDa/nm |
-Macromolecule #1: Actin, alpha skeletal muscle, intermediate form
| Macromolecule | Name: Actin, alpha skeletal muscle, intermediate form / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.387227 KDa |
| Sequence | String: TTALVCDNGS GLVKAGFAGD DAPRAVFPSI VGRPRHQGVM VGMGQKDSYV GDEAQSKRGI LTLKYPIE(HIC)G IITNWD DME KIWHHTFYNE LRVAPEEHPT LLTEAPLNPK ANREKMTQIM FETFNVPAMY VAIQAVLSLY ASGRTTGIVL DSGDGVT HN VPIYEGYALP ...String: TTALVCDNGS GLVKAGFAGD DAPRAVFPSI VGRPRHQGVM VGMGQKDSYV GDEAQSKRGI LTLKYPIE(HIC)G IITNWD DME KIWHHTFYNE LRVAPEEHPT LLTEAPLNPK ANREKMTQIM FETFNVPAMY VAIQAVLSLY ASGRTTGIVL DSGDGVT HN VPIYEGYALP HAIMRLDLAG RDLTDYLMKI LTERGYSFVT TAEREIVRDI KEKLCYVALD FENEMATAAS SSSLEKSY E LPDGQVITIG NERFRCPETL FQPSFIGMES AGIHETTYNS IMKCDIDIRK DLYANNVMSG GTTMYPGIAD RMQKEITAL APSTMKIKII APPERKYSVW IGGSILASLS TFQQMWITKQ EYDEAGPSIV HRKCF |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Details: 10 mM imidazole pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 0.01% NaN3 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K |
| Details | chicken cytoskeletal actin purified from chicken breast muscle |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Average exposure time: 10.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)