[English] 日本語
 Yorodumi
Yorodumi- EMDB-23959: Native RhopH complex of the malaria parasite Plasmodium falciparum -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23959 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native RhopH complex of the malaria parasite Plasmodium falciparum | |||||||||||||||||||||||||||||||||
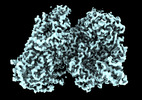
 Map data Map data | CryoEM density map of RhopH complex. | |||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | malaria / rhoptry / complex / soluble / MEMBRANE PROTEIN | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationrhoptry / adhesion of symbiont to microvasculature / symbiont-containing vacuole membrane / cytoplasmic vesicle / host cell cytoplasm / host cell plasma membrane / membrane Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.72 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Ho CM / Jih J | |||||||||||||||||||||||||||||||||
| Funding support |  United States, 10 items United States, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Native structure of the RhopH complex, a key determinant of malaria parasite nutrient acquisition. Authors: Chi-Min Ho / Jonathan Jih / Mason Lai / Xiaorun Li / Daniel E Goldberg / Josh R Beck / Z Hong Zhou /  Abstract: The RhopH complex is implicated in malaria parasites' ability to invade and create new permeability pathways in host erythrocytes, but its mechanisms remain poorly understood. Here, we enrich the ...The RhopH complex is implicated in malaria parasites' ability to invade and create new permeability pathways in host erythrocytes, but its mechanisms remain poorly understood. Here, we enrich the endogenous RhopH complex in a native soluble form, comprising RhopH2, CLAG3.1, and RhopH3, directly from parasite cell lysates and determine its atomic structure using cryo-electron microscopy (cryo-EM), mass spectrometry, and the cryoID program. CLAG3.1 is positioned between RhopH2 and RhopH3, which both share substantial binding interfaces with CLAG3.1 but make minimal contacts with each other. The forces stabilizing individual subunits include 13 intramolecular disulfide bonds. Notably, CLAG3.1 residues 1210 to 1223, previously predicted to constitute a transmembrane helix, are embedded within a helical bundle formed by residues 979 to 1289 near the C terminus of CLAG3.1. Buried in the core of the RhopH complex and largely shielded from solvent, insertion of this putative transmembrane helix into the erythrocyte membrane would likely require a large conformational rearrangement. Given the unusually high disulfide content of the complex, it is possible that such a rearrangement could be initiated by the breakage of allosteric disulfide bonds, potentially triggered by interactions at the erythrocyte membrane. This first direct observation of an exported transmembrane protein-in a soluble, trafficking state and with atomic details of buried putative membrane-insertion helices-offers insights into the assembly and trafficking of RhopH and other parasite-derived complexes to the erythrocyte membrane. Our study demonstrates the potential the endogenous structural proteomics approach holds for elucidating the molecular mechanisms of hard-to-isolate complexes in their native, functional forms. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23959.map.gz emd_23959.map.gz | 7.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23959-v30.xml emd-23959-v30.xml emd-23959.xml emd-23959.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23959.png emd_23959.png | 256.3 KB | ||
| Filedesc metadata |  emd-23959.cif.gz emd-23959.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23959 http://ftp.pdbj.org/pub/emdb/structures/EMD-23959 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23959 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23959 | HTTPS FTP |
-Related structure data
| Related structure data |  7mrwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23959.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23959.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM density map of RhopH complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : High molecular mass rhoptry protein complex (RhopH complex)
| Entire | Name: High molecular mass rhoptry protein complex (RhopH complex) |
|---|---|
| Components |
|
-Supramolecule #1: High molecular mass rhoptry protein complex (RhopH complex)
| Supramolecule | Name: High molecular mass rhoptry protein complex (RhopH complex) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cytoadherence linked asexual protein 3.1
| Macromolecule | Name: Cytoadherence linked asexual protein 3.1 / type: protein_or_peptide / ID: 1 Details: CLAG3.1, subunit of the soluble form of the RhopH complex Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 167.444219 KDa |
| Sequence | String: MVSFFKTPIF ILIIFLYLNE KVICSINENQ NENDTISQNV NQHENINQNV NDNDNIEQLK SMIGNDELHK NLTILEKLIL ESLEKDKLK YPLLKQGTEQ LIDISKFNKK NITDADDETY IIPTVQSTFH DIVKYEHLIK EQSIEIYNSD ISDKIKKKIF I VRTLKTIK ...String: MVSFFKTPIF ILIIFLYLNE KVICSINENQ NENDTISQNV NQHENINQNV NDNDNIEQLK SMIGNDELHK NLTILEKLIL ESLEKDKLK YPLLKQGTEQ LIDISKFNKK NITDADDETY IIPTVQSTFH DIVKYEHLIK EQSIEIYNSD ISDKIKKKIF I VRTLKTIK LMLIPLNSYK QNNDLKSALE ELNNVFTNKE AQEESSPIGD HGTFFRKLLT HVRTIKENED IENKGETLIL GD NKIDVMN SNDFFFTTNS NVKFMENLDD ITNQYGLGLI NHLGPHLIAL GHFTVLKLAL KNYKNYFEAK SIKFFSWQKI LEF SMSDRF KVLDMMCDHE SVYYSEKKRR KTYLKVDRSN TSMECNILEY LLHYFNKYQL EIIKTTQDTD FDLHGMMEHK YIKD YFFSF MCNDPKECII YHTNQFKKEA NEENTFPEQE EPNRQISAFN LYLNYYYFMK RYSSYGVKKT LYVHLLNLTG LLNYD TRAY VTSLYLPGYY NAVEMSFTEE KEFSKLFESL IQCIEKCHSD QARQISKDSN LLNNITKCDL CKGAFLYANM KFDEVP SML QKFYVYLTKG LKIQKVSSLI KTLDIYQDYS NYLSHDINWY TFLFLFRLTS FKEIAKKNVA EAMYLNIKDE DTFNKTV VT NYWYPSPIKK YYTLYVRKHI PNNLVDELEK LMKSGTLEKM KKSLTFLVHV NSFLQLDFFH QLNEPPLGLP RSYPLSLV L EHKFKEWMNS SPAGFYFSNY QNPYIRKDLH DKVLSQKFEP PKMNQWNKVL KSLIECAYDM YFEQRHVKNL YKYHNIYNI NNKLMLMRDS IDLYKNNFDD VLFFADIFNM RKYMTATPVY KKVKDRVYHT LHSITGNSVN FYKYGIIYGF KVNKEILKEV VDELYSIYN FNTDIFTDTS FLQTVYLLFR RIEETYRTQR RDDKISVNNV FFMNVANNYS KLNKEEREIE IHNSMASRYY A KTMFAAFQ MLFSTMLSNN VDNLDKAYGL SENIQVATST SAFLTFAYVY NGSIMDSVTN SLLPPYAKKP ITQLKYGKTF VF SNYFMLA SKMYDMLNYK NLSLLCEYQA VASANFYSAK KVGQFLGRKF LPITTYFLVM RISWTHAFTT GQHLISAFGS PSS TANGKS NASGYKSPES FFFTHGLAAE ASKYLFFYFF TNLYLDAYKS FPGGFGPAIK EQTQHVQEQT YERKPSVHSF NRNF FMELV NGFMYAFCFF AISQMYAYFE NINFYITSNF RFLDRYYGVF NKYFINYAII KLKEITSDLL IKYEREAYLS MKKYG YLGE VIAARLSPKD KIMNYVHETN EDIMSNLRRY DMENAFKNKM STYVDDFAFF DDCGKNEQFL NERCDYCPVI EEVEET QLF TTTGDKNTNK TTEIKKQTST YIDTEKMNEA DSADSDDEKD SDTPDDELMI SRFH UniProtKB: Cytoadherence linked asexual protein 3.1 |
-Macromolecule #2: High molecular weight rhoptry protein 2
| Macromolecule | Name: High molecular weight rhoptry protein 2 / type: protein_or_peptide / ID: 2 Details: RhopH2, subunit of the soluble form of the RhopH complex Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 162.886344 KDa |
| Sequence | String: MIKVTIFLLL SIFSFNLYGL ELNEKVSIKY GAEQGVGSAD SNTKLCSDIL KYLYMDEYLS EGDKATFEKK CHNVIGNIRN TFSNKNTIK EGNEFLMSIL HMKSLYGNNN NNNAGSESDV TLKSLYLSLK GSQNTEGESE VPSDDEINKT IMNFVKFNKY L LDNSNDIK ...String: MIKVTIFLLL SIFSFNLYGL ELNEKVSIKY GAEQGVGSAD SNTKLCSDIL KYLYMDEYLS EGDKATFEKK CHNVIGNIRN TFSNKNTIK EGNEFLMSIL HMKSLYGNNN NNNAGSESDV TLKSLYLSLK GSQNTEGESE VPSDDEINKT IMNFVKFNKY L LDNSNDIK KVHDFLVLTS QSNENLLPNK EKLFEQIVDQ IKYFDEYFFA SGGKIKVKKG YLKYNFLDIY KQPVCSAYLH LC SRYYESV SIYIRLKKVF NGIPAFLDKN CRKVKGEEFK KLMDMELKHN HIVERFDKYI ISDDLYYVNM KVFDLKNVDK IQV SKIDDI NNLNIYEHKE TMHLSAKNLS RYIDIKKELN DEKAYKQLMS AIRKYVTTLT KADSDITYFV KQLDDEEIER FLID LNFFL YNGFLRITED KHLINADDVS PSYINLYRSN NIVALYILKT QYEENKLSEY RAHKFYRRKR VSNITNDMIK KDFTQ TNAL TNLPNLDNKK TTEYYLKEYE NFVENFQPDL HDIMKLQLFF TMAFKDCNVN QNFTETSKKL WFDLLYAYDK FGWFYI HPN EVINSINKTD FVRHVLVSRN FLLKNNDQLT FLETQVAKIV EIINLSLEVD KSPDSLDFSI PMNFFNHKNG YHVMNDD KL KLLTSYEYID SIANNYFFLS EYKNDVFRTG NNFKLYFNLP NIYSLAYQLF NELAININVI TNVPLKKYLK YNASYAYF T LMNMIGKNHD IYSKGSRFVY ASYILGLVFF IESHIDIARL KPKDFFFMKQ SLPIIDHVYH KDLKTLKKNC TLLTDFMKI NKNSQNYSLT HTEEMIKILG LLTVTLWAKE GKKSVYYDDD VSLYRKLMVS CVFNGGETIQ EKLANNIEKS CDISQYGIKS KNLKDMIDI NLSIHKWNPA EIEKLAYSFV LSCKMQKLMY KPMNVEKLPL EDYYKLSLAP DMVKTYHCYK LGKQAAELLE S IILKKKFV RFRVTDAIDV YDFFYIKKVL SSRIKKEYNE FLQDKRAFEK KELETILNNS PFSEEQTMKL INSYECHWFT SY ENFRILW MHASSNLGTG TYLKNFFSEL WQNIRFLFKS KLKIRDMEYF SGDISQMNLL DYYSPMVHSE SHCQEKMQVL FIT LRDSKE ENRSEIAQKV KSAYYQCKLD YYKNHHSDFI HRIHPNDFLN NKVYVLKQPY YLMSNVPLNN PKKVSRLFVT EGTL EYLLL DKINIPECFG PCTKLHFNKV VIKESKQRIY DMTINNALVP EIQPYNRRKY MTIYINEAYI KNIVSDALTS EEIKR HDIQ KGNIKICMGK STYLTEPILT EEHFNLTHKP VYDFSSVKHN LKVFHMKNEH LVSEDPNDDC FINYPLATIN LDISDP YKE ISEDLIKNLY ILKSS UniProtKB: High molecular weight rhoptry protein 2 |
-Macromolecule #3: High molecular weight rhoptry protein 3
| Macromolecule | Name: High molecular weight rhoptry protein 3 / type: protein_or_peptide / ID: 3 Details: RhopH3, subunit of the soluble form of the RhopH complex Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 104.997406 KDa |
| Sequence | String: MRSKHLVTLF IITFLSFSTV KVWGKDVFAG FVTKKLKTLL DCNFALYYNF KGNGPDAGSF LDFVDEPEQF YWFVEHFLSV KFRVPKHLK DKNIHNFTPC LNRSWVSEFL KEYEEPFVNP VMKFLDKEQR LFFTYNFGDV EPQGKYTYFP VKEFHKYCIL P PLIKTNIK ...String: MRSKHLVTLF IITFLSFSTV KVWGKDVFAG FVTKKLKTLL DCNFALYYNF KGNGPDAGSF LDFVDEPEQF YWFVEHFLSV KFRVPKHLK DKNIHNFTPC LNRSWVSEFL KEYEEPFVNP VMKFLDKEQR LFFTYNFGDV EPQGKYTYFP VKEFHKYCIL P PLIKTNIK DGESGEFLKY QLNKEEYKVF LSSVGSQMTA IKNLYSTVED EQRKQLLKVI IENESTNDIS VQCPTYNIKL HY TKECANS NNILKCIDEF LRKTCEKKTE SKHPSADLCE HLQFLFESLK NPYLDNFKKF MTNSDFTLIK PQSVWNVPIF DIY KPKNYL DSVQNLDTEC FKKLNSKNLI FLSFHDDIPN NPYYNVELQE IVKLSTYTYS IFDKLYNFFF VFKKSGAPIS PVSV KELSH NITDFSFKED NSEIQCQNVR KSLDLEVDVE TMKGIAAEKL CKIIEKFILT KDDASKPEKS DIHRGFRILC ILIST HVEA YNIVRQLLNM ESMISLTRYT SLYIHKFFKS VTLLKGNFLY KNNKAIRYSR ACSKASLHVP SVLYRRNIYI PETFLS LYL GLSNLVSSNP SSPFFEYAII EFLVTYYNKG SEKFVLYFIS IISVLYINEY YYEQLSCFYP KEFELIKSRM IHPNIVD RI LKGIDNLMKS TRYDKMRTMY LDFESSDIFS REKVFTALYN FDSFIKTNEQ LKKKNLEEIS EIPVQLETSN DGIGYRKQ D VLYETDKPQT MDEASYEETV DEDAHHVNEK QHSAHFLDAI AEKDILEEKT KDQDLEIELY KYMGPLKEQS KSTSAASTS DEISGSEGPS TESTSTGNQG EDKTTDNTYK EMEELEEAEG TSNLKKGLEF YKSSLKLDQL DKEKPKKKKS KRKKKRDSSS DRILLEESK TFTSENEL UniProtKB: High molecular weight rhoptry protein 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7mrw: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















