+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23803 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A. baumannii MsbA in complex with TBT1 decoupler | |||||||||
 Map data Map data | A. baumannii MsbA in complex with TBT1 decoupler | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationATPase-coupled lipid transmembrane transporter activity / ABC-type transporter activity / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.97 Å | |||||||||
 Authors Authors | Thelot F / Liao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Distinct allosteric mechanisms of first-generation MsbA inhibitors. Authors: François A Thélot / Wenyi Zhang / KangKang Song / Chen Xu / Jing Huang / Maofu Liao /   Abstract: ATP-binding cassette (ABC) transporters couple adenosine 5′-triphosphate (ATP) hydrolysis to substrate transport across biological membranes. Although many are promising drug targets, their ...ATP-binding cassette (ABC) transporters couple adenosine 5′-triphosphate (ATP) hydrolysis to substrate transport across biological membranes. Although many are promising drug targets, their mechanisms of modulation by small-molecule inhibitors remain largely unknown. Two first-generation inhibitors of the MsbA transporter, tetrahydrobenzothiophene 1 (TBT1) and G247, induce opposite effects on ATP hydrolysis. Using single-particle cryo–electron microscopy and functional assays, we show that TBT1 and G247 bind adjacent yet separate pockets in the MsbA transmembrane domains. Two TBT1 molecules asymmetrically occupy the substrate-binding site, which leads to a collapsed inward-facing conformation with decreased distance between the nucleotide-binding domains (NBDs). By contrast, two G247 molecules symmetrically increase NBD distance in a wide inward-open state of MsbA. The divergent mechanisms of action of these MsbA inhibitors provide important insights into ABC transporter pharmacology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23803.map.gz emd_23803.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23803-v30.xml emd-23803-v30.xml emd-23803.xml emd-23803.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23803.png emd_23803.png | 79.5 KB | ||
| Filedesc metadata |  emd-23803.cif.gz emd-23803.cif.gz | 5.5 KB | ||
| Others |  emd_23803_additional_1.map.gz emd_23803_additional_1.map.gz | 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23803 http://ftp.pdbj.org/pub/emdb/structures/EMD-23803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23803 | HTTPS FTP |
-Validation report
| Summary document |  emd_23803_validation.pdf.gz emd_23803_validation.pdf.gz | 414.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23803_full_validation.pdf.gz emd_23803_full_validation.pdf.gz | 414.1 KB | Display | |
| Data in XML |  emd_23803_validation.xml.gz emd_23803_validation.xml.gz | 4.5 KB | Display | |
| Data in CIF |  emd_23803_validation.cif.gz emd_23803_validation.cif.gz | 5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23803 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23803 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23803 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23803 | HTTPS FTP |
-Related structure data
| Related structure data |  7metMC  7mewC  7ritC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10778 (Title: A. baumannii MsbA in complex with TBT1 / Data size: 267.1 EMPIAR-10778 (Title: A. baumannii MsbA in complex with TBT1 / Data size: 267.1 Data #1: motion-corrected micrographs of A. baumannii MsbA in complex with TBT1 stimulator [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23803.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23803.map.gz / Format: CCP4 / Size: 6.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A. baumannii MsbA in complex with TBT1 decoupler | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
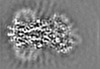
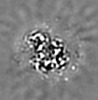
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unfiltered, unsharpened map
| File | emd_23803_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered, unsharpened map | ||||||||||||
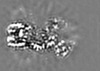
| Projections & Slices |
| ||||||||||||
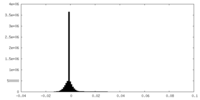
| Density Histograms |
- Sample components
Sample components
-Entire : A. baumannii MsbA in complex with TBT1 decoupler
| Entire | Name: A. baumannii MsbA in complex with TBT1 decoupler |
|---|---|
| Components |
|
-Supramolecule #1: A. baumannii MsbA in complex with TBT1 decoupler
| Supramolecule | Name: A. baumannii MsbA in complex with TBT1 decoupler / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 130 KDa |
-Macromolecule #1: ATP-dependent lipid A-core flippase
| Macromolecule | Name: ATP-dependent lipid A-core flippase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: ABC-type lipid A-core oligosaccharide transporter |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii (bacteria) Acinetobacter baumannii (bacteria) |
| Molecular weight | Theoretical: 67.758688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MMHHHHHHHH GGVNQDFKVY LRLISYLKPY WGVALLVLIG FGMNSATEVS VAKLIKFIID AIQNASRAD LDWFPLLIIL LVFFRGLGLF MGGYYTAVIS RSLVFSIRQE VYAKLLRLPA QYYLDNSSGH ITAKIMYNVE Q LTAASSES ...String: MGSSHHHHHH SSGLVPRGSH MMHHHHHHHH GGVNQDFKVY LRLISYLKPY WGVALLVLIG FGMNSATEVS VAKLIKFIID AIQNASRAD LDWFPLLIIL LVFFRGLGLF MGGYYTAVIS RSLVFSIRQE VYAKLLRLPA QYYLDNSSGH ITAKIMYNVE Q LTAASSES LKTIVRDGMI TLGLLGYLFY TNWRLTICIM VFLPIIGILV RKASKRMRKL SMQVQDTMGD VNHVVQESIN GN AVVKSFA GEESEQERFY KSSEENLKRG LKMVIVQNLN SPVVQVVMAC AMALIVWLAL RPQILGNTTA GEFVAYITAA GLL SKPVKN LTDVNEKLQR GLAAAHSVFE LLDLPEEQNS GELKPQLQGA IRFDHVVLNY ADGTQAIKDF SLDIRPGETV ALVG RSGAG KTSLVNMLVR FQEVSSGQIY LDDLPIRDIE LSSLRTQIAM VNQQVVLFNR TVRENIAYGQ LHNASDEDVI AAAKA AYAH DFIMNLPNGY DTVLGAQGLN LSGGQRQRIA IARAILKNAP ILILDEATSA LDNESEHFIQ QAFDEAMQDR TTIVIA HRL STIENADRIV VMDRGQIVEQ GTHQELLAKH GAYYQLHQRN FEDH UniProtKB: Lipid A export permease/ATP-binding protein MsbA |
-Macromolecule #2: 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carbo...
| Macromolecule | Name: 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxylic acid type: ligand / ID: 2 / Number of copies: 2 / Formula: Z5G |
|---|---|
| Molecular weight | Theoretical: 335.805 Da |
| Chemical component information | 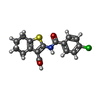 ChemComp-Z5G: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.97 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 67714 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)