[English] 日本語
 Yorodumi
Yorodumi- EMDB-23792: CryoEM structure of monoclonal Fab 045-09 2B05 binding the latera... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23792 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
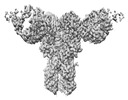
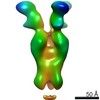
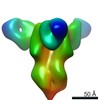
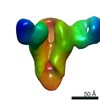
| Title | CryoEM structure of monoclonal Fab 045-09 2B05 binding the lateral patch of influenza virus H1 HA | |||||||||
 Map data Map data | CryoEM map of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hemagglutinin / monoclonal antibody / influenza virus / IMMUNE SYSTEM / Viral protein-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Influenza A virus (strain swl A/California/04/2009 H1N1) Influenza A virus (strain swl A/California/04/2009 H1N1) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Han J / Ward A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2021 Journal: Sci Transl Med / Year: 2021Title: First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Authors: Jenna J Guthmiller / Julianna Han / Lei Li / Alec W Freyn / Sean T H Liu / Olivia Stovicek / Christopher T Stamper / Haley L Dugan / Micah E Tepora / Henry A Utset / Dalia J Bitar / Natalie ...Authors: Jenna J Guthmiller / Julianna Han / Lei Li / Alec W Freyn / Sean T H Liu / Olivia Stovicek / Christopher T Stamper / Haley L Dugan / Micah E Tepora / Henry A Utset / Dalia J Bitar / Natalie J Hamel / Siriruk Changrob / Nai-Ying Zheng / Min Huang / Florian Krammer / Raffael Nachbagauer / Peter Palese / Andrew B Ward / Patrick C Wilson /  Abstract: Broadly neutralizing antibodies are critical for protection against both drifted and shifted influenza viruses. Here, we reveal that first exposure to the 2009 pandemic H1N1 influenza virus recalls ...Broadly neutralizing antibodies are critical for protection against both drifted and shifted influenza viruses. Here, we reveal that first exposure to the 2009 pandemic H1N1 influenza virus recalls memory B cells that are specific to the conserved receptor-binding site (RBS) or lateral patch epitopes of the hemagglutinin (HA) head domain. Monoclonal antibodies (mAbs) generated against these epitopes are broadly neutralizing against H1N1 viruses spanning 40 years of viral evolution and provide potent protection in vivo. Lateral patch-targeting antibodies demonstrated near universal binding to H1 viruses, and RBS-binding antibodies commonly cross-reacted with H3N2 viruses and influenza B viruses. Lateral patch-targeting mAbs were restricted to expressing the variable heavy-chain gene VH3-23 with or without the variable kappa-chain gene VK1-33 and often had a Y-x-R motif within the heavy-chain complementarity determining region 3 to make key contacts with HA. Moreover, lateral patch antibodies that used both VH3-23 and VK1-33 maintained neutralizing capability with recent pH1N1 strains that acquired mutations near the lateral patch. RBS-binding mAbs used a diverse repertoire but targeted the RBS epitope similarly and made extensive contacts with the major antigenic site Sb. Together, our data indicate that RBS- and lateral patch-targeting clones are abundant within the human memory B cell pool, and universal vaccine strategies should aim to drive antibodies against both conserved head and stalk epitopes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23792.map.gz emd_23792.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23792-v30.xml emd-23792-v30.xml emd-23792.xml emd-23792.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23792_fsc.xml emd_23792_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_23792.png emd_23792.png | 84.2 KB | ||
| Masks |  emd_23792_msk_1.map emd_23792_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23792.cif.gz emd-23792.cif.gz | 6.6 KB | ||
| Others |  emd_23792_half_map_1.map.gz emd_23792_half_map_1.map.gz emd_23792_half_map_2.map.gz emd_23792_half_map_2.map.gz | 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23792 http://ftp.pdbj.org/pub/emdb/structures/EMD-23792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23792 | HTTPS FTP |
-Related structure data
| Related structure data |  7memMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23792.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23792.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23792_msk_1.map emd_23792_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM half map of monoclonal Fab 045-09 2B05...
| File | emd_23792_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM half map of monoclonal Fab 045-09 2B05...
| File | emd_23792_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of monoclonal Fab 045-09 2B05 binding the latera...
| Entire | Name: CryoEM structure of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of monoclonal Fab 045-09 2B05 binding the latera...
| Supramolecule | Name: CryoEM structure of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: monoclonal Fab 045-09 2B05
| Supramolecule | Name: monoclonal Fab 045-09 2B05 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Hemagglutinin
| Supramolecule | Name: Hemagglutinin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1, #4 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (strain swl A/California/04/2009 H1N1) Influenza A virus (strain swl A/California/04/2009 H1N1) |
-Macromolecule #1: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (strain swl A/California/04/2009 H1N1) Influenza A virus (strain swl A/California/04/2009 H1N1)Strain: swl A/California/04/2009 H1N1 |
| Molecular weight | Theoretical: 36.729402 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADPGDTLCIG YHANNSTDTV DTVLEKNVTV THSVNLLEDK HNGKLCKLRG VAPLHLGKCN IAGWILGNPE CESLSTASSW SYIVETPSS DNGTCYPGDF IDYEELREQL SSVSSFERFE IFPKTSSWPN HDSNKGVTAA CPHAGAKSFY KNLIWLVKKG N SYPKLSKS ...String: ADPGDTLCIG YHANNSTDTV DTVLEKNVTV THSVNLLEDK HNGKLCKLRG VAPLHLGKCN IAGWILGNPE CESLSTASSW SYIVETPSS DNGTCYPGDF IDYEELREQL SSVSSFERFE IFPKTSSWPN HDSNKGVTAA CPHAGAKSFY KNLIWLVKKG N SYPKLSKS YINDKGKEVL VLWGIHHPST SADQQSLYQN ADTYVFVGSS RYSKKFKPEI AIRPKVRDQE GRMNYYWTLV EP GDKITFE ATGNLVVPRY AFAMERNAGS GIIISDTPVH DCNTTCQTPK GAINTSLPFQ NIHPITIGKC PKYVKSTKLR LAT GLRNIP SIQSR UniProtKB: Hemagglutinin |
-Macromolecule #2: Light chain of monoclonal antibody 045-09 2B05
| Macromolecule | Name: Light chain of monoclonal antibody 045-09 2B05 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Cell: B cell Homo sapiens (human) / Cell: B cell |
| Molecular weight | Theoretical: 11.76912 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSAFVGDRVT IACQASQDIR IHLNWYQQKP GKAPKLLIYD ASNLEAGVPS RFSGSGSGTD FTFTISSLQP EDIATYYCQ HYHNLPRTFG GGTKVEIK |
-Macromolecule #3: Heavy chain of monoclonal antibody 045-09 2B05
| Macromolecule | Name: Heavy chain of monoclonal antibody 045-09 2B05 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Cell: B cell Homo sapiens (human) / Cell: B cell |
| Molecular weight | Theoretical: 12.805238 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VQLLESGGGL VQPGGSLSLS CAASGFTFSS FAMSWVRQAP VKGLEWVSMI SAGGGNTYYA DSVKGRFTIS RDNSKSTLYL QMSSLTAED TAVYYCAKSD SSGFQYGRRE FWGQGTLVTV S |
-Macromolecule #4: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (strain swl A/California/04/2009 H1N1) Influenza A virus (strain swl A/California/04/2009 H1N1)Strain: swl A/California/04/2009 H1N1 |
| Molecular weight | Theoretical: 19.904998 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADLKS TQNAIDGITN KVNSVIEKMN TQFTAVGKEF NHLEKRIENL NKKVDDGFL DIWTYNAELL VLLENERTLD YHDSNVKNLY EKVRSQLKNN AKEIGNGCFE FYHKCDNTCM ESVKNGTYDY P KYSEEAKL NREEID UniProtKB: Hemagglutinin |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.75 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: TBS + LMNG |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 49.27 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)