[English] 日本語
 Yorodumi
Yorodumi- EMDB-23544: Human phosphofructokinase-1 liver type bound to activator NA-11 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23544 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human phosphofructokinase-1 liver type bound to activator NA-11 | |||||||||
 Map data Map data | Human phosphofructokinase-1 liver type bound to activator NA-11 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycolysis / phosphofructokinase / liver type / activated / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information6-phosphofructokinase complex / 6-phosphofructokinase / fructose binding / 6-phosphofructokinase activity / fructose-6-phosphate binding / fructose 1,6-bisphosphate metabolic process / fructose 6-phosphate metabolic process / canonical glycolysis / Glycolysis / response to glucose ...6-phosphofructokinase complex / 6-phosphofructokinase / fructose binding / 6-phosphofructokinase activity / fructose-6-phosphate binding / fructose 1,6-bisphosphate metabolic process / fructose 6-phosphate metabolic process / canonical glycolysis / Glycolysis / response to glucose / glycolytic process / negative regulation of insulin secretion / kinase binding / secretory granule lumen / ficolin-1-rich granule lumen / Neutrophil degranulation / extracellular exosome / extracellular region / ATP binding / metal ion binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Lynch EM / Kollman JM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Selective activation of PFKL suppresses the phagocytic oxidative burst. Authors: Neri Amara / Madison P Cooper / Maria A Voronkova / Bradley A Webb / Eric M Lynch / Justin M Kollman / Taylur Ma / Kebing Yu / Zijuan Lai / Dewakar Sangaraju / Nobuhiko Kayagaki / Kim Newton ...Authors: Neri Amara / Madison P Cooper / Maria A Voronkova / Bradley A Webb / Eric M Lynch / Justin M Kollman / Taylur Ma / Kebing Yu / Zijuan Lai / Dewakar Sangaraju / Nobuhiko Kayagaki / Kim Newton / Matthew Bogyo / Steven T Staben / Vishva M Dixit /  Abstract: In neutrophils, nicotinamide adenine dinucleotide phosphate (NADPH) generated via the pentose phosphate pathway fuels NADPH oxidase NOX2 to produce reactive oxygen species for killing invading ...In neutrophils, nicotinamide adenine dinucleotide phosphate (NADPH) generated via the pentose phosphate pathway fuels NADPH oxidase NOX2 to produce reactive oxygen species for killing invading pathogens. However, excessive NOX2 activity can exacerbate inflammation, as in acute respiratory distress syndrome (ARDS). Here, we use two unbiased chemical proteomic strategies to show that small-molecule LDC7559, or a more potent designed analog NA-11, inhibits the NOX2-dependent oxidative burst in neutrophils by activating the glycolytic enzyme phosphofructokinase-1 liver type (PFKL) and dampening flux through the pentose phosphate pathway. Accordingly, neutrophils treated with NA-11 had reduced NOX2-dependent outputs, including neutrophil cell death (NETosis) and tissue damage. A high-resolution structure of PFKL confirmed binding of NA-11 to the AMP/ADP allosteric activation site and explained why NA-11 failed to agonize phosphofructokinase-1 platelet type (PFKP) or muscle type (PFKM). Thus, NA-11 represents a tool for selective activation of PFKL, the main phosphofructokinase-1 isoform expressed in immune cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23544.map.gz emd_23544.map.gz | 11 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23544-v30.xml emd-23544-v30.xml emd-23544.xml emd-23544.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23544.png emd_23544.png | 238.3 KB | ||
| Filedesc metadata |  emd-23544.cif.gz emd-23544.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23544 http://ftp.pdbj.org/pub/emdb/structures/EMD-23544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23544 | HTTPS FTP |
-Validation report
| Summary document |  emd_23544_validation.pdf.gz emd_23544_validation.pdf.gz | 384.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23544_full_validation.pdf.gz emd_23544_full_validation.pdf.gz | 384.1 KB | Display | |
| Data in XML |  emd_23544_validation.xml.gz emd_23544_validation.xml.gz | 6.6 KB | Display | |
| Data in CIF |  emd_23544_validation.cif.gz emd_23544_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23544 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23544 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23544 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23544 | HTTPS FTP |
-Related structure data
| Related structure data |  7lw1MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23544.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23544.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human phosphofructokinase-1 liver type bound to activator NA-11 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human phosphofructokinase-1 liver type bound to activator NA-11
| Entire | Name: Human phosphofructokinase-1 liver type bound to activator NA-11 |
|---|---|
| Components |
|
-Supramolecule #1: Human phosphofructokinase-1 liver type bound to activator NA-11
| Supramolecule | Name: Human phosphofructokinase-1 liver type bound to activator NA-11 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATP-dependent 6-phosphofructokinase, liver type
| Macromolecule | Name: ATP-dependent 6-phosphofructokinase, liver type / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: 6-phosphofructokinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85.120375 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MAAVDLEKLR ASGAGKAIGV LTSGGDAQGM NAAVRAVTRM GIYVGAKVFL IYEGYEGLVE GGENIKQANW LSVSNIIQLG GTIIGSARC KAFTTREGRR AAAYNLVQHG ITNLCVIGGD GSLTGANIFR SEWGSLLEEL VAEGKISETT ARTYSHLNIA G LVGSIDND ...String: MAAVDLEKLR ASGAGKAIGV LTSGGDAQGM NAAVRAVTRM GIYVGAKVFL IYEGYEGLVE GGENIKQANW LSVSNIIQLG GTIIGSARC KAFTTREGRR AAAYNLVQHG ITNLCVIGGD GSLTGANIFR SEWGSLLEEL VAEGKISETT ARTYSHLNIA G LVGSIDND FCGTDMTIGT DSALHRIMEV IDAITTTAQS HQRTFVLEVM GRHCGYLALV SALASGADWL FIPEAPPEDG WE NFMCERL GETRSRGSRL NIIIIAEGAI DRNGKPISSS YVKDLVVQRL GFDTRVTVLG HVQRGGTPSA FDRILSSKMG MEA VMALLE ATPDTPACVV TLSGNQSVRL PLMECVQMTK EVQKAMDDKR FDEATQLRGG SFENNWNIYK LLAHQKPPKE KSNF SLAIL NVGAPAAGMN AAVRSAVRTG ISHGHTVYVV HDGFEGLAKG QVQEVGWHDV AGWLGRGGSM LGTKRTLPKG QLESI VENI RIYGIHALLV VGGFEAYEGV LQLVEARGRY EELCIVMCVI PATISNNVPG TDFSLGSDTA VNAAMESCDR IKQSAS GTK RRVFIVETMG GYCGYLATVT GIAVGADAAY VFEDPFNIHD LKVNVEHMTE KMKTDIQRGL VLRNEKCHDY YTTEFLY NL YSSEGKGVFD CRTNVLGHLQ QGGAPTPFDR NYGTKLGVKA MLWLSEKLRE VYRKGRVFAN APDSACVIGL KKKAVAFS P VTELKKDTDF EHRMPREQWW LSLRLMLKML AQYRISMAAY VSGELEHVTR RTLSMDKGF UniProtKB: ATP-dependent 6-phosphofructokinase, liver type |
-Macromolecule #2: N-{(11S)-2-[2-(5-hydroxypent-1-yn-1-yl)phenyl]-4H,10H-pyrazolo[5,...
| Macromolecule | Name: N-{(11S)-2-[2-(5-hydroxypent-1-yn-1-yl)phenyl]-4H,10H-pyrazolo[5,1-c][1,4]benzoxazepin-7-yl}acetamide type: ligand / ID: 2 / Number of copies: 4 / Formula: YG1 |
|---|---|
| Molecular weight | Theoretical: 401.458 Da |
| Chemical component information |  ChemComp-YG1: |
-Macromolecule #3: 1,6-di-O-phosphono-beta-D-fructofuranose
| Macromolecule | Name: 1,6-di-O-phosphono-beta-D-fructofuranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: FBP |
|---|---|
| Molecular weight | Theoretical: 340.116 Da |
| Chemical component information | 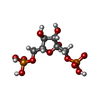 ChemComp-FBP: |
-Macromolecule #4: 6-O-phosphono-beta-D-fructofuranose
| Macromolecule | Name: 6-O-phosphono-beta-D-fructofuranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: F6P |
|---|---|
| Molecular weight | Theoretical: 260.136 Da |
| Chemical component information |  ChemComp-F6P: |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 4 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 2529 / Average exposure time: 10.0 sec. / Average electron dose: 90.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)





















