+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2335 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The capsid of mycobacteriophage Araucaria | |||||||||
 Map data Map data | Icosahedral Reconstruction of the Araucaria capsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mycobacteriophage / Araucaria / single-particle / Mycobacterium abscessus subsp. bolletii / Siphoviridae / electron microscopy / mycobacteria. | |||||||||
| Biological species | Mycobacteriophage Araucaria | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 30.0 Å | |||||||||
 Authors Authors | Sassi M / Bebeacua C / Drancourt M / Cambillau C | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: The first structure of a mycobacteriophage, the Mycobacterium abscessus subsp. bolletii phage Araucaria. Authors: Mohamed Sassi / Cecilia Bebeacua / Michel Drancourt / Christian Cambillau /  Abstract: The unique characteristics of the waxy mycobacterial cell wall raise questions about specific structural features of their bacteriophages. No structure of any mycobacteriophage is available, although ...The unique characteristics of the waxy mycobacterial cell wall raise questions about specific structural features of their bacteriophages. No structure of any mycobacteriophage is available, although ∼3,500 have been described to date. To fill this gap, we embarked in a genomic and structural study of a bacteriophage from Mycobacterium abscessus subsp. bolletii, a member of the Mycobacterium abscessus group. This opportunistic pathogen is responsible for respiratory tract infections in patients with lung disorders, particularly cystic fibrosis. M. abscessus subsp. bolletii was isolated from respiratory tract specimens, and bacteriophages were observed in the cultures. We report here the genome annotation and characterization of the M. abscessus subsp. bolletii prophage Araucaria, as well as the first single-particle electron microscopy reconstruction of the whole virion. Araucaria belongs to Siphoviridae and possesses a 64-kb genome containing 89 open reading frames (ORFs), among which 27 could be annotated with certainty. Although its capsid and connector share close similarity with those of several phages from Gram-negative (Gram(-)) or Gram(+) bacteria, its most distinctive characteristic is the helical tail decorated by radial spikes, possibly host adhesion devices, according to which the phage name was chosen. Its host adsorption device, at the tail tip, assembles features observed in phages binding to protein receptors, such as phage SPP1. All together, these results suggest that Araucaria may infect its mycobacterial host using a mechanism involving adhesion to cell wall saccharides and protein, a feature that remains to be further explored. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2335.map.gz emd_2335.map.gz | 22.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2335-v30.xml emd-2335-v30.xml emd-2335.xml emd-2335.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2335.tif EMD-2335.tif | 49.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2335 http://ftp.pdbj.org/pub/emdb/structures/EMD-2335 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2335 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2335 | HTTPS FTP |
-Validation report
| Summary document |  emd_2335_validation.pdf.gz emd_2335_validation.pdf.gz | 188.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2335_full_validation.pdf.gz emd_2335_full_validation.pdf.gz | 187.7 KB | Display | |
| Data in XML |  emd_2335_validation.xml.gz emd_2335_validation.xml.gz | 5.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2335 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2335 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2335 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2335 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2335.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2335.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral Reconstruction of the Araucaria capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.95 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
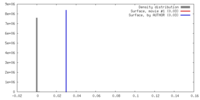
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mycobacteriophage Araucaria Capsid
| Entire | Name: Mycobacteriophage Araucaria Capsid |
|---|---|
| Components |
|
-Supramolecule #1000: Mycobacteriophage Araucaria Capsid
| Supramolecule | Name: Mycobacteriophage Araucaria Capsid / type: sample / ID: 1000 Details: Capsid particles were selected from a sample containing whole phages. Oligomeric state: Icosahedral / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 2.8 MDa / Theoretical: 2.8 MDa / Method: Approximation with Chimera |
-Supramolecule #1: Mycobacteriophage Araucaria
| Supramolecule | Name: Mycobacteriophage Araucaria / type: virus / ID: 1 / Name.synonym: Araucaria / Sci species name: Mycobacteriophage Araucaria / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No / Syn species name: Araucaria |
|---|---|
| Host (natural) | Organism:  Mycobacterium abscessus subsp. bolletii (bacteria) Mycobacterium abscessus subsp. bolletii (bacteria)Strain: CIP108541T / synonym: BACTERIA(EUBACTERIA) |
| Virus shell | Shell ID: 1 / Name: Capsid T7 / Diameter: 600 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: PBS Buffer |
|---|---|
| Staining | Type: NEGATIVE Details: Grids with adsorbed viral particles were stained with 2% uranyl acetate for 20 seconds. |
| Grid | Details: 300 copper mesh grids with a film of colodion and carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 150,000 times magnification |
| Date | Jul 1, 2012 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD (2k x 2k) / Number real images: 1500 / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 48500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 48500 |
| Sample stage | Specimen holder: Room temperature holder / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Details | Particles were processed using a Maximum Likelihood approach imposing ICOSAHEDRAL symmetry. |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 30.0 Å / Resolution method: OTHER / Software - Name: EMAN2, Spider, Xmipp / Number images used: 7431 |
| Final angle assignment | Details: ICOSAHEDRAL |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | 60 copies of the HK97 MCP hexamer were manually fitted and automatically refined. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)