[English] 日本語
 Yorodumi
Yorodumi- EMDB-5003: Icosahedral structure of bacteriophage epsilon15 at 4.5 Angstrom ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5003 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Icosahedral structure of bacteriophage epsilon15 at 4.5 Angstrom resolution | |||||||||
 Map data Map data | Icosahedral reconstruction of bacteriophage Epsilon15 at 4.5 Angstrom resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / virus / epsilon15 / de novo model / gp7 / gp10 / icosahedral | |||||||||
| Function / homology | : / Major coat protein-like / : / Major capsid protein GP7 / viral capsid, decoration / viral capsid / Major coat protein / Major capsid protein Function and homology information Function and homology information | |||||||||
| Biological species |   epsilon15 (virus) epsilon15 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Jiang W / Baker ML / Jakana J / Weigele PR / King J / Chiu W | |||||||||
 Citation Citation |  Journal: Nature / Year: 2008 Journal: Nature / Year: 2008Title: Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Authors: Wen Jiang / Matthew L Baker / Joanita Jakana / Peter R Weigele / Jonathan King / Wah Chiu /  Abstract: A half-century after the determination of the first three-dimensional crystal structure of a protein, more than 40,000 structures ranging from single polypeptides to large assemblies have been ...A half-century after the determination of the first three-dimensional crystal structure of a protein, more than 40,000 structures ranging from single polypeptides to large assemblies have been reported. The challenge for crystallographers, however, remains the growing of a diffracting crystal. Here we report the 4.5-A resolution structure of a 22-MDa macromolecular assembly, the capsid of the infectious epsilon15 (epsilon15) particle, by single-particle electron cryomicroscopy. From this density map we constructed a complete backbone trace of its major capsid protein, gene product 7 (gp7). The structure reveals a similar protein architecture to that of other tailed double-stranded DNA viruses, even in the absence of detectable sequence similarity. However, the connectivity of the secondary structure elements (topology) in gp7 is unique. Protruding densities are observed around the two-fold axes that cannot be accounted for by gp7. A subsequent proteomic analysis of the whole virus identifies these densities as gp10, a 12-kDa protein. Its structure, location and high binding affinity to the capsid indicate that the gp10 dimer functions as a molecular staple between neighbouring capsomeres to ensure the particle's stability. Beyond epsilon15, this method potentially offers a new approach for modelling the backbone conformations of the protein subunits in other macromolecular assemblies at near-native solution states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5003.map.gz emd_5003.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5003-v30.xml emd-5003-v30.xml emd-5003.xml emd-5003.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5003_1.png emd_5003_1.png | 532.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5003 http://ftp.pdbj.org/pub/emdb/structures/EMD-5003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5003 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5003 | HTTPS FTP |
-Related structure data
| Related structure data |  3j40M  3c5b M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5003.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5003.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral reconstruction of bacteriophage Epsilon15 at 4.5 Angstrom resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
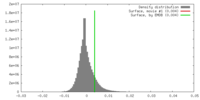
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : bacteriophage epsilon15
| Entire | Name: bacteriophage epsilon15 |
|---|---|
| Components |
|
-Supramolecule #1000: bacteriophage epsilon15
| Supramolecule | Name: bacteriophage epsilon15 / type: sample / ID: 1000 / Oligomeric state: icosahedral / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 22 MDa |
-Supramolecule #1: epsilon15
| Supramolecule | Name: epsilon15 / type: virus / ID: 1 / Name.synonym: epsilon15 / Sci species name: epsilon15 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: epsilon15 |
|---|---|
| Host (natural) | Organism:  Salmonella enterica subsp. enterica serovar Anatum (bacteria) Salmonella enterica subsp. enterica serovar Anatum (bacteria)synonym: BACTERIA(EUBACTERIA) |
| Molecular weight | Experimental: 22 MDa |
| Virus shell | Shell ID: 1 / Diameter: 700 Å / T number (triangulation number): 7 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 50 mM Tris pH7.5, 25 mM NaCl and 10 mM MgCl2 |
|---|---|
| Grid | Details: Quantifoil R2/2 grid |
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER / Details: Vitrification instrument: Vitrobot |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3000SFF |
|---|---|
| Temperature | Average: 4.2 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 1235 / Average electron dose: 25 e/Å2 / Bits/pixel: 14 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.6 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Topentry / Specimen holder model: GATAN HELIUM |
- Image processing
Image processing
| CTF correction | Details: per particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN / Number images used: 36259 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)