[English] 日本語
 Yorodumi
Yorodumi- EMDB-23207: Structure and function at the lipid-protein interface of a pentam... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23207 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and function at the lipid-protein interface of a pentameric ligand-gated ion channel | |||||||||
 Map data Map data | Unliganded ELIC in SMA-nanodiscs of E. Coli inner Membrane | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pentameric Ligand-gated Ion Channels / Nanodisc / Cys-loop receptor / Styrene-maleic acid / Membrane Protein / Protein-lipid interface | |||||||||
| Function / homology |  Function and homology information Function and homology informationextracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Dickeya dadantii (bacteria) / Dickeya dadantii (bacteria) /  Dickeya dadantii (strain 3937) (bacteria) Dickeya dadantii (strain 3937) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Grosman C / Kumar P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structure and function at the lipid-protein interface of a pentameric ligand-gated ion channel. Authors: Pramod Kumar / Gisela D Cymes / Claudio Grosman /  Abstract: Although it has long been proposed that membrane proteins may contain tightly bound lipids, their identity, the structure of their binding sites, and their functional and structural relevance have ...Although it has long been proposed that membrane proteins may contain tightly bound lipids, their identity, the structure of their binding sites, and their functional and structural relevance have remained elusive. To some extent, this is because tightly bound lipids are often located at the periphery of proteins, where the quality of density maps is usually poorer, and because they may be outcompeted by detergent molecules used during standard purification procedures. As a step toward characterizing natively bound lipids in the superfamily of pentameric ligand-gated ion channels (pLGICs), we applied single-particle cryogenic electron microscopy to fragments of native membrane obtained in the complete absence of detergent-solubilization steps. Because of the heterogeneous lipid composition of membranes in the secretory pathway of eukaryotic cells, we chose to study a bacterial pLGIC (ELIC) expressed in 's inner membrane. We obtained a three-dimensional reconstruction of unliganded ELIC (2.5-Å resolution) that shows clear evidence for two types of tightly bound lipid at the protein-bulk-membrane interface. One of them was consistent with a "regular" diacylated phospholipid, in the cytoplasmic leaflet, whereas the other one was consistent with the tetra-acylated structure of cardiolipin, in the periplasmic leaflet. Upon reconstitution in polar-lipid bilayers, ELIC retained the functional properties characteristic of members of this superfamily, and thus, the fitted atomic model is expected to represent the (long-debated) unliganded-closed, "resting" conformation of this ion channel. Notably, the addition of cardiolipin to phosphatidylcholine membranes restored the ion-channel activity that is largely lost in phosphatidylcholine-only bilayers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23207.map.gz emd_23207.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23207-v30.xml emd-23207-v30.xml emd-23207.xml emd-23207.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23207.png emd_23207.png | 77.4 KB | ||
| Masks |  emd_23207_msk_1.map emd_23207_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23207.cif.gz emd-23207.cif.gz | 5.9 KB | ||
| Others |  emd_23207_half_map_1.map.gz emd_23207_half_map_1.map.gz emd_23207_half_map_2.map.gz emd_23207_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23207 http://ftp.pdbj.org/pub/emdb/structures/EMD-23207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23207 | HTTPS FTP |
-Validation report
| Summary document |  emd_23207_validation.pdf.gz emd_23207_validation.pdf.gz | 704.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23207_full_validation.pdf.gz emd_23207_full_validation.pdf.gz | 704.3 KB | Display | |
| Data in XML |  emd_23207_validation.xml.gz emd_23207_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_23207_validation.cif.gz emd_23207_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23207 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23207 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23207 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23207 | HTTPS FTP |
-Related structure data
| Related structure data |  7l6qMC  7l6uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23207.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23207.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unliganded ELIC in SMA-nanodiscs of E. Coli inner Membrane | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.073 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23207_msk_1.map emd_23207_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
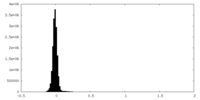
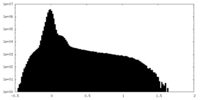
| Density Histograms |
-Half map: Half-map 1
| File | emd_23207_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
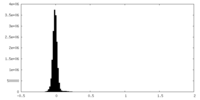
| Density Histograms |
-Half map: Half-map 2
| File | emd_23207_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
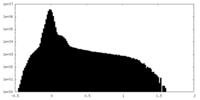
| Density Histograms |
- Sample components
Sample components
-Entire : Unliganded Erwinia chrysanthemi ligand gated ion channel in styre...
| Entire | Name: Unliganded Erwinia chrysanthemi ligand gated ion channel in styrene-maleic-acid nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: Unliganded Erwinia chrysanthemi ligand gated ion channel in styre...
| Supramolecule | Name: Unliganded Erwinia chrysanthemi ligand gated ion channel in styrene-maleic-acid nanodiscs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Dickeya dadantii (bacteria) Dickeya dadantii (bacteria) |
| Molecular weight | Theoretical: 184.2 kDa/nm |
-Macromolecule #1: Gamma-aminobutyric-acid receptor subunit beta-1
| Macromolecule | Name: Gamma-aminobutyric-acid receptor subunit beta-1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dickeya dadantii (strain 3937) (bacteria) / Strain: 3937 Dickeya dadantii (strain 3937) (bacteria) / Strain: 3937 |
| Molecular weight | Theoretical: 36.879 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APADNAADAR PVDVSVSIFI NKIYGVNTLE QTYKVDGYIV AQWTGKPRKT PGDKPLIVEN TQIERWINNG LWVPALEFIN VVGSPDTGN KRLMLFPDGR VIYNARFLGS FSNDMDFRLF PFDRQQFVLE LEPFSYNNQQ LRFSDIQVYT ENIDNEEIDE W WIRGKAST ...String: APADNAADAR PVDVSVSIFI NKIYGVNTLE QTYKVDGYIV AQWTGKPRKT PGDKPLIVEN TQIERWINNG LWVPALEFIN VVGSPDTGN KRLMLFPDGR VIYNARFLGS FSNDMDFRLF PFDRQQFVLE LEPFSYNNQQ LRFSDIQVYT ENIDNEEIDE W WIRGKAST HISDIRYDHL SSVQPNQNEF SRITVRIDAV RNPSYYLWSF ILPLGLIIAA SWSVFWLESF SERLQTSFTL ML TVVAYAF YTSNILPRLP YTTVIDQMII AGYGSIFAAI LLIIFAHHRQ ANGVEDDLLI QRCRLAFPLG FLAIGCVLVI RGI TL UniProtKB: Gamma-aminobutyric-acid receptor subunit beta-1 |
-Macromolecule #2: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]o...
| Macromolecule | Name: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate type: ligand / ID: 2 / Number of copies: 5 / Formula: PGW |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
-Macromolecule #3: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 3 / Number of copies: 5 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Material: GOLD / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 56.23 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1239532 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)