+Search query
-Structure paper
| Title | Structure and function at the lipid-protein interface of a pentameric ligand-gated ion channel. |
|---|---|
| Journal, issue, pages | Proc Natl Acad Sci U S A, Vol. 118, Issue 23, Year 2021 |
| Publish date | Jun 8, 2021 |
 Authors Authors | Pramod Kumar / Gisela D Cymes / Claudio Grosman /  |
| PubMed Abstract | Although it has long been proposed that membrane proteins may contain tightly bound lipids, their identity, the structure of their binding sites, and their functional and structural relevance have ...Although it has long been proposed that membrane proteins may contain tightly bound lipids, their identity, the structure of their binding sites, and their functional and structural relevance have remained elusive. To some extent, this is because tightly bound lipids are often located at the periphery of proteins, where the quality of density maps is usually poorer, and because they may be outcompeted by detergent molecules used during standard purification procedures. As a step toward characterizing natively bound lipids in the superfamily of pentameric ligand-gated ion channels (pLGICs), we applied single-particle cryogenic electron microscopy to fragments of native membrane obtained in the complete absence of detergent-solubilization steps. Because of the heterogeneous lipid composition of membranes in the secretory pathway of eukaryotic cells, we chose to study a bacterial pLGIC (ELIC) expressed in 's inner membrane. We obtained a three-dimensional reconstruction of unliganded ELIC (2.5-Å resolution) that shows clear evidence for two types of tightly bound lipid at the protein-bulk-membrane interface. One of them was consistent with a "regular" diacylated phospholipid, in the cytoplasmic leaflet, whereas the other one was consistent with the tetra-acylated structure of cardiolipin, in the periplasmic leaflet. Upon reconstitution in polar-lipid bilayers, ELIC retained the functional properties characteristic of members of this superfamily, and thus, the fitted atomic model is expected to represent the (long-debated) unliganded-closed, "resting" conformation of this ion channel. Notably, the addition of cardiolipin to phosphatidylcholine membranes restored the ion-channel activity that is largely lost in phosphatidylcholine-only bilayers. |
 External links External links |  Proc Natl Acad Sci U S A / Proc Natl Acad Sci U S A /  PubMed:34083441 / PubMed:34083441 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.5 - 3.3 Å |
| Structure data | EMDB-23207: Structure and function at the lipid-protein interface of a pentameric ligand-gated ion channel EMDB-23208, PDB-7l6u: |
| Chemicals | 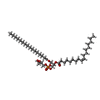 ChemComp-PGW:  ChemComp-CDL: |
| Source |
|
 Keywords Keywords | MEMBRANE PROTEIN / Pentameric Ligand-gated Ion Channels / Nanodisc / Cys-loop receptor / Styrene-maleic acid / Protein-lipid interface / Styrene Maleic Acid Copolymer |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 Dickeya dadantii (bacteria)
Dickeya dadantii (bacteria)