[English] 日本語
 Yorodumi
Yorodumi- EMDB-22497: SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody F... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22497 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / COVID-19 / spike glycoprotein / fusion protein / neutralizing antibodies / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Park YJ / Tortorici MA / Walls AC / Czudnochowski N / Snell G / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Authors: Luca Piccoli / Young-Jun Park / M Alejandra Tortorici / Nadine Czudnochowski / Alexandra C Walls / Martina Beltramello / Chiara Silacci-Fregni / Dora Pinto / Laura E Rosen / John E Bowen / ...Authors: Luca Piccoli / Young-Jun Park / M Alejandra Tortorici / Nadine Czudnochowski / Alexandra C Walls / Martina Beltramello / Chiara Silacci-Fregni / Dora Pinto / Laura E Rosen / John E Bowen / Oliver J Acton / Stefano Jaconi / Barbara Guarino / Andrea Minola / Fabrizia Zatta / Nicole Sprugasci / Jessica Bassi / Alessia Peter / Anna De Marco / Jay C Nix / Federico Mele / Sandra Jovic / Blanca Fernandez Rodriguez / Sneha V Gupta / Feng Jin / Giovanni Piumatti / Giorgia Lo Presti / Alessandra Franzetti Pellanda / Maira Biggiogero / Maciej Tarkowski / Matteo S Pizzuto / Elisabetta Cameroni / Colin Havenar-Daughton / Megan Smithey / David Hong / Valentino Lepori / Emiliano Albanese / Alessandro Ceschi / Enos Bernasconi / Luigia Elzi / Paolo Ferrari / Christian Garzoni / Agostino Riva / Gyorgy Snell / Federica Sallusto / Katja Fink / Herbert W Virgin / Antonio Lanzavecchia / Davide Corti / David Veesler /      Abstract: Analysis of the specificity and kinetics of neutralizing antibodies (nAbs) elicited by SARS-CoV-2 infection is crucial for understanding immune protection and identifying targets for vaccine design. ...Analysis of the specificity and kinetics of neutralizing antibodies (nAbs) elicited by SARS-CoV-2 infection is crucial for understanding immune protection and identifying targets for vaccine design. In a cohort of 647 SARS-CoV-2-infected subjects, we found that both the magnitude of Ab responses to SARS-CoV-2 spike (S) and nucleoprotein and nAb titers correlate with clinical scores. The receptor-binding domain (RBD) is immunodominant and the target of 90% of the neutralizing activity present in SARS-CoV-2 immune sera. Whereas overall RBD-specific serum IgG titers waned with a half-life of 49 days, nAb titers and avidity increased over time for some individuals, consistent with affinity maturation. We structurally defined an RBD antigenic map and serologically quantified serum Abs specific for distinct RBD epitopes leading to the identification of two major receptor-binding motif antigenic sites. Our results explain the immunodominance of the receptor-binding motif and will guide the design of COVID-19 vaccines and therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22497.map.gz emd_22497.map.gz | 561.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22497-v30.xml emd-22497-v30.xml emd-22497.xml emd-22497.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22497.png emd_22497.png | 33.5 KB | ||
| Filedesc metadata |  emd-22497.cif.gz emd-22497.cif.gz | 7.3 KB | ||
| Others |  emd_22497_additional_1.map.gz emd_22497_additional_1.map.gz emd_22497_half_map_1.map.gz emd_22497_half_map_1.map.gz emd_22497_half_map_2.map.gz emd_22497_half_map_2.map.gz | 122.3 MB 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22497 http://ftp.pdbj.org/pub/emdb/structures/EMD-22497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22497 | HTTPS FTP |
-Related structure data
| Related structure data |  7jvaMC  7jv2C  7jv4C  7jv6C  7jvcC  7jw0C  7jx3C  7jxcC  7jxdC  7jxeC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22497.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22497.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_22497_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
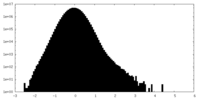
| Density Histograms |
-Half map: Half map A
| File | emd_22497_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
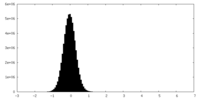
| Density Histograms |
-Half map: Half map B
| File | emd_22497_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
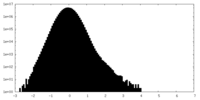
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody F...
| Entire | Name: SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody F...
| Supramolecule | Name: SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: spike glycoprotein
| Supramolecule | Name: spike glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: S2A4 neutralizing antibody Fab fragment
| Supramolecule | Name: S2A4 neutralizing antibody Fab fragment / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: S2A4 Fab heavy chain
| Macromolecule | Name: S2A4 Fab heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.302878 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFTFS SYWMNWVRQA PGKGLEWVAN IKQDGSEKYY VDSVKGRFTI SRDNAKNSLF LQMNSLRAE DTAVYYCARV WWLRGSFDYW GQGTLVTVSS |
-Macromolecule #2: S2A4 Fab light chain
| Macromolecule | Name: S2A4 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.845768 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NFMLTQPHSV SESPGKTVTI SCTGSSGSIA SNYVQWYQQR PGSAPTTVIY EDNQRPSGVP DRFSGSIDSS SNSASLTISG LKTEDEADY YCQSYDSSNH VVFGGGTKLT VL |
-Macromolecule #3: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.532797 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF L GVYYHKNN ...String: MGILPSPGMP ALLSLVSLLS VLLMGCVAET GTQCVNLTTR TQLPPAYTNS FTRGVYYPDK VFRSSVLHST QDLFLPFFSN VTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF L GVYYHKNN KSWMESEFRV YSSANNCTFE YVSQPFLMDL EGKQGNFKNL REFVFKNIDG YFKIYSKHTP INLVRDLPQG FS ALEPLVD LPIGINITRF QTLLALHRSY LTPGDSSSGW TAGAAAYYVG YLQPRTFLLK YNENGTITDA VDCALDPLSE TKC TLKSFT VEKGIYQTSN FRVQPTESIV RFPNITNLCP FGEVFNATRF ASVYAWNRKR ISNCVADYSV LYNSASFSTF KCYG VSPTK LNDLCFTNVY ADSFVIRGDE VRQIAPGQTG KIADYNYKLP DDFTGCVIAW NSNNLDSKVG GNYNYLYRLF RKSNL KPFE RDISTEIYQA GSTPCNGVEG FNCYFPLQSY GFQPTNGVGY QPYRVVVLSF ELLHAPATVC GPKKSTNLVK NKCVNF NFN GLTGTGVLTE SNKKFLPFQQ FGRDIADTTD AVRDPQTLEI LDITPCSFGG VSVITPGTNT SNQVAVLYQD VNCTEVP VA IHADQLTPTW RVYSTGSNVF QTRAGCLIGA EHVNNSYECD IPIGAGICAS YQTQTNSPSG AGSVASQSII AYTMSLGA E NSVAYSNNSI AIPTNFTISV TTEILPVSMT KTSVDCTMYI CGDSTECSNL LLQYGSFCTQ LNRALTGIAV EQDKNTQEV FAQVKQIYKT PPIKDFGGFN FSQILPDPSK PSKRSFIEDL LFNKVTLADA GFIKQYGDCL GDIAARDLIC AQKFNGLTVL PPLLTDEMI AQYTSALLAG TITSGWTFGA GAALQIPFAM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS T ASALGKLQ DVVNQNAQAL NTLVKQLSSN FGAISSVLND ILSRLDPPEA EVQIDRLITG RLQSLQTYVT QQLIRAAEIR AS ANLAATK MSECVLGQSK RVDFCGKGYH LMSFPQSAPH GVVFLHVTYV PAQEKNFTTA PAICHDGKAH FPREGVFVSN GTH WFVTQR NFYEPQIITT DNTFVSGNCD VVIGIVNNTV YDPLQPELDS FKEELDKYFK NHTSPDVDLG DISGINASVV NIQK EIDRL NEVAKNLNES LIDLQELGKY EQYIKGSGRE NLYFQGGGGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGHH HHHHH H UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 64141 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)