[English] 日本語
 Yorodumi
Yorodumi- EMDB-22417: ORC-O1AAA: Human Origin Recognition Complex (ORC) with dynamic/un... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22417 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | ORC-O1AAA: Human Origin Recognition Complex (ORC) with dynamic/unresolved ORC2 WH | ||||||||||||
 Map data Map data | combination of global and focused refinement utilizing the 'vop max' function in chimera | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | replication / AAA+ / ORC / DNA-binding | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpolar body extrusion after meiotic divisions / CDC6 association with the ORC:origin complex / origin recognition complex / E2F-enabled inhibition of pre-replication complex formation / nuclear origin of replication recognition complex / nuclear pre-replicative complex / inner kinetochore / DNA replication preinitiation complex / mitotic DNA replication checkpoint signaling / neural precursor cell proliferation ...polar body extrusion after meiotic divisions / CDC6 association with the ORC:origin complex / origin recognition complex / E2F-enabled inhibition of pre-replication complex formation / nuclear origin of replication recognition complex / nuclear pre-replicative complex / inner kinetochore / DNA replication preinitiation complex / mitotic DNA replication checkpoint signaling / neural precursor cell proliferation / G1/S-Specific Transcription / regulation of DNA replication / DNA replication origin binding / protein polymerization / Activation of the pre-replicative complex / DNA replication initiation / Activation of ATR in response to replication stress / glial cell proliferation / heterochromatin / Assembly of the ORC complex at the origin of replication / Assembly of the pre-replicative complex / Orc1 removal from chromatin / DNA replication / chromosome, telomeric region / nuclear body / nucleotide binding / chromatin binding / centrosome / chromatin / nucleolus / negative regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / nucleoplasm / ATP binding / metal ion binding / membrane / nucleus / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Jaremko MJ / Joshua-Tor L | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: The dynamic nature of the human origin recognition complex revealed through five cryoEM structures. Authors: Matt J Jaremko / Kin Fan On / Dennis R Thomas / Bruce Stillman / Leemor Joshua-Tor /  Abstract: Genome replication is initiated from specific origin sites established by dynamic events. The Origin Recognition Complex (ORC) is necessary for orchestrating the initiation process by binding to ...Genome replication is initiated from specific origin sites established by dynamic events. The Origin Recognition Complex (ORC) is necessary for orchestrating the initiation process by binding to origin DNA, recruiting CDC6, and assembling the MCM replicative helicase on DNA. Here we report five cryoEM structures of the human ORC (HsORC) that illustrate the native flexibility of the complex. The absence of ORC1 revealed a compact, stable complex of ORC2-5. Introduction of ORC1 opens the complex into several dynamic conformations. Two structures revealed dynamic movements of the ORC1 AAA+ and ORC2 winged-helix domains that likely impact DNA incorporation into the ORC core. Additional twist and pinch motions were observed in an open ORC conformation revealing a hinge at the ORC5·ORC3 interface that may facilitate ORC binding to DNA. Finally, a structure of ORC was determined with endogenous DNA bound in the core revealing important differences between human and yeast origin recognition. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22417.map.gz emd_22417.map.gz | 115.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22417-v30.xml emd-22417-v30.xml emd-22417.xml emd-22417.xml | 29.2 KB 29.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22417.png emd_22417.png | 156.5 KB | ||
| Filedesc metadata |  emd-22417.cif.gz emd-22417.cif.gz | 8.4 KB | ||
| Others |  emd_22417_additional_1.map.gz emd_22417_additional_1.map.gz emd_22417_additional_2.map.gz emd_22417_additional_2.map.gz | 117 MB 116.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22417 http://ftp.pdbj.org/pub/emdb/structures/EMD-22417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22417 | HTTPS FTP |
-Related structure data
| Related structure data |  7jpoMC  7jppC  7jpqC  7jprC  7jpsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22417.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22417.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | combination of global and focused refinement utilizing the 'vop max' function in chimera | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
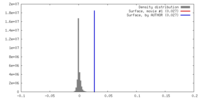
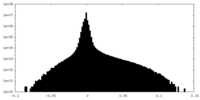
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: global refinement
| File | emd_22417_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | global refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
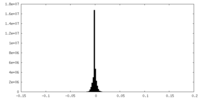
| Density Histograms |
-Additional map: focused refinement
| File | emd_22417_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
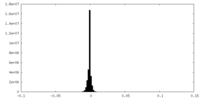
| Density Histograms |
- Sample components
Sample components
-Entire : ORC-O1AAA
| Entire | Name: ORC-O1AAA |
|---|---|
| Components |
|
-Supramolecule #1: ORC-O1AAA
| Supramolecule | Name: ORC-O1AAA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: 5 subunit complex with the ORC2 winged-helix domain in a dynamic/unresolved state |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 296.53 KDa |
-Macromolecule #1: Origin recognition complex subunit 1
| Macromolecule | Name: Origin recognition complex subunit 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.310887 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPQIRSRSL AAQEPASVLE EARLRLHVSA VPESLPCREQ EFQDIYNFVE SKLLDHTGGC MYISGVPGTG KTATVHEVIR CLQQAAQAN DVPPFQYIEV NGMKLTEPHQ VYVQILQKLT GQKATANHAA ELLAKQFCTR GSPQETTVLL VDELDLLWTH K QDIMYNLF ...String: MAPQIRSRSL AAQEPASVLE EARLRLHVSA VPESLPCREQ EFQDIYNFVE SKLLDHTGGC MYISGVPGTG KTATVHEVIR CLQQAAQAN DVPPFQYIEV NGMKLTEPHQ VYVQILQKLT GQKATANHAA ELLAKQFCTR GSPQETTVLL VDELDLLWTH K QDIMYNLF DWPTHKEARL VVLAIANTMD LPERIMMNRV SSRLGLTRMC FQPYTYSQLQ QILRSRLKHL KAFEDDAIQL VA RKVAALS GDARRCLDIC RRATEICEFS QQKPDSPGLV TIAHSMEAVD EMFSSSYITA IKNSSVLEQS FLRAILAEFR RSG LEEATF QQIYSQHVAL CRMEGLPYPT MSETMAVCSH LGSCRLLLVE PSRNDLLLRV RLNVSQDDVL YALKDE UniProtKB: Origin recognition complex subunit 1 |
-Macromolecule #2: Origin recognition complex subunit 2
| Macromolecule | Name: Origin recognition complex subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.063375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKPELKEDK MLEVHFVGDD DVLNHILDRE GGAKLKKERA QLLVNPKKII KKPEYDLEED DQEVLKDQNY VEIMGRDVQE SLKNGSATG GGNKVYSFQN RKHSEKMAKL ASELAKTPQK SVSFSLKNDP EITINVPQSS KGHSASDKVQ PKNNDKSEFL S TAPRSLRK ...String: MSKPELKEDK MLEVHFVGDD DVLNHILDRE GGAKLKKERA QLLVNPKKII KKPEYDLEED DQEVLKDQNY VEIMGRDVQE SLKNGSATG GGNKVYSFQN RKHSEKMAKL ASELAKTPQK SVSFSLKNDP EITINVPQSS KGHSASDKVQ PKNNDKSEFL S TAPRSLRK RLIVPRSHSD SESEYSASNS EDDEGVAQEH EEDTNAVIFS QKIQAQNRVV SAPVGKETPS KRMKRDKTSD LV EEYFEAH SSSKVLTSDR TLQKLKRAKL DQQTLRNLLS KVSPSFSAEL KQLNQQYEKL FHKWMLQLHL GFNIVLYGLG SKR DLLERF RTTMLQDSIH VVINGFFPGI SVKSVLNSIT EEVLDHMGTF RSILDQLDWI VNKFKEDSSL ELFLLIHNLD SQML RGEKS QQIIGQLSSL HNIYLIASID HLNAPLMWDH AKQSLFNWLW YETTTYSPYT EETSYENSLL VKQSGSLPLS SLTHV LRSL TPNARGIFRL LIKYQLDNQD NPSYIGLSFQ DFYQQCREAF LVNSDLTLRA QLTEFRDHKL IRTKKGTDGV EYLLIP VDN GTLTDFLEKE EEEA UniProtKB: Origin recognition complex subunit 2 |
-Macromolecule #3: Origin recognition complex subunit 3
| Macromolecule | Name: Origin recognition complex subunit 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.436133 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATSSMSKGC FVFKPNSKKR KISLPIEDYF NKGKNEPEDS KLRFETYQLI WQQMKSENER LQEELNKNLF DNLIEFLQKS HSGFQKNSR DLGGQIKLRE IPTAALVLGV NVTDHDLTFG SLTEALQNNV TPYVVSLQAK DCPDMKHFLQ KLISQLMDCC V DIKSKEEE ...String: MATSSMSKGC FVFKPNSKKR KISLPIEDYF NKGKNEPEDS KLRFETYQLI WQQMKSENER LQEELNKNLF DNLIEFLQKS HSGFQKNSR DLGGQIKLRE IPTAALVLGV NVTDHDLTFG SLTEALQNNV TPYVVSLQAK DCPDMKHFLQ KLISQLMDCC V DIKSKEEE SVHVTQRKTH YSMDSLSSWY MTVTQKTDPK MLSKKRTTSS QWQSPPVVVI LKDMESFATK VLQDFIIISS QH LHEFPLI LIFGIATSPI IIHRLLPHAV SSLLCIELFQ SLSCKEHLTT VLDKLLLTTQ FPFKINEKVL QVLTNIFLYH DFS VQNFIK GLQLSLLEHF YSQPLSVLCC NLPEAKRRIN FLSNNQCENI RRLPSFRRYV EKQASEKQVA LLTNERYLKE ETQL LLENL HVYHMNYFLV LRCLHKFTSS LPKYPLGRQI RELYCTCLEK NIWDSEEYAS VLQLLRMLAK DELMTILEKC FKVFK SYCE NHLGSTAKRI EEFLAQFQSL DAETKEEEDA SGSQPKGLQK TDLYHLQKSL LEMKELRRSK KQTKFEVLRE NVVNFI DCL VREYLLPPET QPLHEVVYFS AAHALREHLN AAPRIALHTA LNNPYYYLKN EALKSEEGCI PNIAPDICIA YKLHLEC SR LINLVDWSEA FATVVTAAEK MDANSATSEE MNEIIHARFI RAVSELELLG FIKPTKQKTD HVARLTWGGC UniProtKB: Origin recognition complex subunit 3 |
-Macromolecule #4: Origin recognition complex subunit 4
| Macromolecule | Name: Origin recognition complex subunit 4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.443266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSRKSKSNS LIHTECLSQV QRILRERFCR QSPHSNLFGV QVQYKHLSEL LKRTALHGES NSVLIIGPRG SGKTMLINHA LKELMEIEE VSENVLQVHL NGLLQINDKI ALKEITRQLN LENVVGDKVF GSFAENLSFL LEALKKGDRT SSCPVIFILD E FDLFAHHK ...String: MSSRKSKSNS LIHTECLSQV QRILRERFCR QSPHSNLFGV QVQYKHLSEL LKRTALHGES NSVLIIGPRG SGKTMLINHA LKELMEIEE VSENVLQVHL NGLLQINDKI ALKEITRQLN LENVVGDKVF GSFAENLSFL LEALKKGDRT SSCPVIFILD E FDLFAHHK NQTLLYNLFD ISQSAQTPIA VIGLTCRLDI LELLEKRVKS RFSHRQIHLM NSFGFPQYVK IFKEQLSLPA EF PDKVFAE KWNENVQYLS EDRSVQEVLQ KHFNISKNLR SLHMLLMLAL NRVTASHPFM TAVDLMEASQ LCSMDSKANI VHG LSVLEI CLIIAMKHLN DIYEEEPFNF QMVYNEFQKF VQRKAHSVYN FEKPVVMKAF EHLQQLELIK PMERTSGNSQ REYQ LMKLL LDNTQIMNAL QKYPNCPTDV RQWATSSLSW L UniProtKB: Origin recognition complex subunit 4 |
-Macromolecule #5: Origin recognition complex subunit 5
| Macromolecule | Name: Origin recognition complex subunit 5 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.298867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPHLENVVLC RESQVSILQS LFGERHHFSF PSIFIYGHTA SGKTYVTQTL LKTLELPHVF VNCVECFTLR LLLEQILNKL NHLSSSEDG CSTEITCETF NDFVRLFKQV TTAENLKDQT VYIVLDKAEY LRDMEANLLP GFLRLQELAD RNVTVLFLSE I VWEKFRPN ...String: MPHLENVVLC RESQVSILQS LFGERHHFSF PSIFIYGHTA SGKTYVTQTL LKTLELPHVF VNCVECFTLR LLLEQILNKL NHLSSSEDG CSTEITCETF NDFVRLFKQV TTAENLKDQT VYIVLDKAEY LRDMEANLLP GFLRLQELAD RNVTVLFLSE I VWEKFRPN TGCFEPFVLY FPDYSIGNLQ KILSHDHPPE YSADFYAAYI NILLGVFYTV CRDLKELRHL AVLNFPKYCE PV VKGEASE RDTRKLWRNI EPHLKKAMQT VYLREISSSQ WEKLQKDDTD PGQLKGLSAH THVELPYYSK FILIAAYLAS YNP ARTDKR FFLKHHGKIK KTNFLKKHEK TSNSLLGPKP FPLDRLLAIL YSIVDSRVAP TANIFSQITS LVTLQLLTLV GHDD QLDGP KYKCTVSLDF IRAIARTVNF DIIKYLYDFL UniProtKB: Origin recognition complex subunit 5 |
-Macromolecule #6: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 3 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #8: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 9068 / Average exposure time: 6.0 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.4 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation Coefficient | ||||||||||||
| Output model |  PDB-7jpo: |
-Atomic model buiding 2
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE | ||||||||||||
| Output model |  PDB-7jpo: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)