[English] 日本語
 Yorodumi
Yorodumi- EMDB-2234: Electron cryo-microscopy of a head-tailed virus infecting extreme... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2234 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of a head-tailed virus infecting extremely halophilic archaea | |||||||||
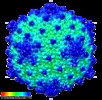
 Map data Map data | Reconstruction of archaeal virus HVTV-1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Icosahedral capsid | |||||||||
| Biological species | HVTV-1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.5 Å | |||||||||
 Authors Authors | Pietila MK / Laurinmaki P / Russell DA / Ko CC / Jacobs-Sera D / Butcher SJ / Bamford DH / Hendrix RW | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2013 Journal: J Virol / Year: 2013Title: Insights into head-tailed viruses infecting extremely halophilic archaea. Authors: Maija K Pietilä / Pasi Laurinmäki / Daniel A Russell / Ching-Chung Ko / Deborah Jacobs-Sera / Sarah J Butcher / Dennis H Bamford / Roger W Hendrix /  Abstract: Extremophilic archaea, both hyperthermophiles and halophiles, dominate in habitats where rather harsh conditions are encountered. Like all other organisms, archaeal cells are susceptible to viral ...Extremophilic archaea, both hyperthermophiles and halophiles, dominate in habitats where rather harsh conditions are encountered. Like all other organisms, archaeal cells are susceptible to viral infections, and to date, about 100 archaeal viruses have been described. Among them, there are extraordinary virion morphologies as well as the common head-tailed viruses. Although approximately half of the isolated archaeal viruses belong to the latter group, no three-dimensional virion structures of these head-tailed viruses are available. Thus, rigorous comparisons with bacteriophages are not yet warranted. In the present study, we determined the genome sequences of two of such viruses of halophiles and solved their capsid structures by cryo-electron microscopy and three-dimensional image reconstruction. We show that these viruses are inactivated, yet remain intact, at low salinity and that their infectivity is regained when high salinity is restored. This enabled us to determine their three-dimensional capsid structures at low salinity to a ∼10-Å resolution. The genetic and structural data showed that both viruses belong to the same T-number class, but one of them has enlarged its capsid to accommodate a larger genome than typically associated with a T=7 capsid by inserting an additional protein into the capsid lattice. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2234.map.gz emd_2234.map.gz | 144.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2234-v30.xml emd-2234-v30.xml emd-2234.xml emd-2234.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2234.tif emd_2234.tif | 961.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2234 http://ftp.pdbj.org/pub/emdb/structures/EMD-2234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2234 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2234 | HTTPS FTP |
-Validation report
| Summary document |  emd_2234_validation.pdf.gz emd_2234_validation.pdf.gz | 290.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2234_full_validation.pdf.gz emd_2234_full_validation.pdf.gz | 289.4 KB | Display | |
| Data in XML |  emd_2234_validation.xml.gz emd_2234_validation.xml.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2234 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2234 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2234 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2234 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2234.map.gz / Format: CCP4 / Size: 589.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2234.map.gz / Format: CCP4 / Size: 589.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of archaeal virus HVTV-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : archaeal virus HVTV-1
| Entire | Name: archaeal virus HVTV-1 |
|---|---|
| Components |
|
-Supramolecule #1000: archaeal virus HVTV-1
| Supramolecule | Name: archaeal virus HVTV-1 / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: HVTV-1
| Supramolecule | Name: HVTV-1 / type: virus / ID: 1 / Sci species name: HVTV-1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Haloarcula vallismortis (Halophile) / synonym: ARCHAEA Haloarcula vallismortis (Halophile) / synonym: ARCHAEA |
| Virus shell | Shell ID: 1 / Diameter: 955 Å / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 / Details: 20mM Tris-HCl |
|---|---|
| Grid | Details: Quantifoil holey carbon film on 300 mesh copper grid. |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Oct 8, 2009 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Bits/pixel: 12 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 62000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 10.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: AUTO3DEM / Number images used: 3530 |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















