[English] 日本語
 Yorodumi
Yorodumi- EMDB-22307: The negative stain EM structure of the human DNA LigIIIalpha-XRCC... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22307 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The negative stain EM structure of the human DNA LigIIIalpha-XRCC1 complex; conformer 2 | |||||||||||||||
 Map data Map data | Refined 3D map of full-length DNA LigaseIII alpha in complex with full-length XRCC1; conformer 2 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | DNA repair / protein-protein interactions / DNA BINDING PROTEIN | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / negative staining / Resolution: 31.5 Å | |||||||||||||||
 Authors Authors | Sverzhinsky A / Pascal JM | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase IIIα within a flexible DNA repair complex. Authors: Michal Hammel / Ishtiaque Rashid / Aleksandr Sverzhinsky / Yasin Pourfarjam / Miaw-Sheue Tsai / Tom Ellenberger / John M Pascal / In-Kwon Kim / John A Tainer / Alan E Tomkinson /   Abstract: The XRCC1-DNA ligase IIIα complex (XL) is critical for DNA single-strand break repair, a key target for PARP inhibitors in cancer cells deficient in homologous recombination. Here, we combined ...The XRCC1-DNA ligase IIIα complex (XL) is critical for DNA single-strand break repair, a key target for PARP inhibitors in cancer cells deficient in homologous recombination. Here, we combined biophysical approaches to gain insights into the shape and conformational flexibility of the XL as well as XRCC1 and DNA ligase IIIα (LigIIIα) alone. Structurally-guided mutational analyses based on the crystal structure of the human BRCT-BRCT heterodimer identified the network of salt bridges that together with the N-terminal extension of the XRCC1 C-terminal BRCT domain constitute the XL molecular interface. Coupling size exclusion chromatography with small angle X-ray scattering and multiangle light scattering (SEC-SAXS-MALS), we determined that the XL is more compact than either XRCC1 or LigIIIα, both of which form transient homodimers and are highly disordered. The reduced disorder and flexibility allowed us to build models of XL particles visualized by negative stain electron microscopy that predict close spatial organization between the LigIIIα catalytic core and both BRCT domains of XRCC1. Together our results identify an atypical BRCT-BRCT interaction as the stable nucleating core of the XL that links the flexible nick sensing and catalytic domains of LigIIIα to other protein partners of the flexible XRCC1 scaffold. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22307.map.gz emd_22307.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22307-v30.xml emd-22307-v30.xml emd-22307.xml emd-22307.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22307.png emd_22307.png | 33 KB | ||
| Filedesc metadata |  emd-22307.cif.gz emd-22307.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22307 http://ftp.pdbj.org/pub/emdb/structures/EMD-22307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22307 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22307 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22307.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22307.map.gz / Format: CCP4 / Size: 2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined 3D map of full-length DNA LigaseIII alpha in complex with full-length XRCC1; conformer 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
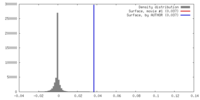
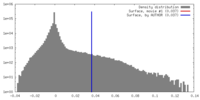
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Full-length DNA LigIII alpha in complex with full-length XRCC1; c...
| Entire | Name: Full-length DNA LigIII alpha in complex with full-length XRCC1; conformer 2 |
|---|---|
| Components |
|
-Supramolecule #1: Full-length DNA LigIII alpha in complex with full-length XRCC1; c...
| Supramolecule | Name: Full-length DNA LigIII alpha in complex with full-length XRCC1; conformer 2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 180 KDa |
-Macromolecule #1: Nuclear DNA ligase III-alpha
| Macromolecule | Name: Nuclear DNA ligase III-alpha / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: DNA ligase (ATP) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MAEQRFCVDY AKRGTAGCKK CKEKIVKGVC RIGKVVPNPF SESGGDMKEW YHIKCMFEKL ERARATTKK IEDLTELEGW EELEDNEKEQ ITQHIADLSS KAAGTPKKKA VVQAKLTTTG Q VTSPVKGA SFVTSTNPRK FSGFSAKPNN SGEAPSSPTP KRSLSSSKCD ...String: MAEQRFCVDY AKRGTAGCKK CKEKIVKGVC RIGKVVPNPF SESGGDMKEW YHIKCMFEKL ERARATTKK IEDLTELEGW EELEDNEKEQ ITQHIADLSS KAAGTPKKKA VVQAKLTTTG Q VTSPVKGA SFVTSTNPRK FSGFSAKPNN SGEAPSSPTP KRSLSSSKCD PRHKDCLLRE FR KLCAMVA DNPSYNTKTQ IIQDFLRKGS AGDGFHGDVY LTVKLLLPGV IKTVYNLNDK QIV KLFSRI FNCNPDDMAR DLEQGDVSET IRVFFEQSKS FPPAAKSLLT IQEVDEFLLR LSKL TKEDE QQQALQDIAS RCTANDLKCI IRLIKHDLKM NSGAKHVLDA LDPNAYEAFK ASRNL QDVV ERVLHNAQEV EKEPGQRRAL SVQASLMTPV QPMLAEACKS VEYAMKKCPN GMFSEI KYD GERVQVHKNG DHFSYFSRSL KPVLPHKVAH FKDYIPQAFP GGHSMILDSE VLLIDNK TG KPLPFGTLGV HKKAAFQDAN VCLFVFDCIY FNDVSLMDRP LCERRKFLHD NMVEIPNR I MFSEMKRVTK ALDLADMITR VIQEGLEGLV LKDVKGTYEP GKRHWLKVKK DYLNEGAMA DTADLVVLGA FYGQGSKGGM MSIFLMGCYD PGSQKWCTVT KCAGGHDDAT LARLQNELDM VKISKDPSK IPSWLKVNKI YYPDFIVPDP KKAAVWEITG AEFSKSEAHT ADGISIRFPR C TRIRDDKD WKSATNLPQL KELYQLSKEK ADFTVVAGDE GSSTTGGSSE ENKGPSGSAV SR KAPSKPS ASTKKAEGKL SNSNSKDGNM QTAKPSAMKV GEKLATKSSP VKVGEKRKAA DET LCQTKV LLDIFTGVRL YLPPSTPDFS RLRRYFVAFD GDLVQEFDMT SATHVLGSRD KNPA AQQVS PEWIWACIRK RRLVAPCWSH PQFEK |
-Macromolecule #2: DNA repair protein XRCC1
| Macromolecule | Name: DNA repair protein XRCC1 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHMPEI RLRHVVSCSS QDSTHCAENL LKADTYRKWR AAKAGEKTIS VVLQLEKEEQ IHSVDI GND GSAFVEVLVG SSAGGAGEQD YEVLLVTSSF MSPSESRSGS NPNRVRMFGP DKLVRAA AE KRWDRVKIVC SQPYSKDSPF GLSFVRFHSP PDKDEAEAPS ...String: HHHHHHMPEI RLRHVVSCSS QDSTHCAENL LKADTYRKWR AAKAGEKTIS VVLQLEKEEQ IHSVDI GND GSAFVEVLVG SSAGGAGEQD YEVLLVTSSF MSPSESRSGS NPNRVRMFGP DKLVRAA AE KRWDRVKIVC SQPYSKDSPF GLSFVRFHSP PDKDEAEAPS QKVTVTKLGQ FRVKEEDE S ANSLRPGALF FSRINKTSPV TASDPAGPSY AAATLQASSA ASSASPVSRA IGSTSKPQE SPKGKRKLDL NQEEKKTPSK PPAQLSPSVP KRPKLPAPTR TPATAPVPAR AQGAVTGKPR GEGTEPRRP RAGPEELGKI LQGVVVVLSG FQNPFRSELR DKALELGAKY RPDWTRDSTH L ICAFANTP KYSQVLGLGG RIVRKEWVLD CHRMRRRLPS RRYLMAGPGS SSEEDEASHS GG SGDEAPK LPQKQPQTKT KPTQAAGPSS PQKPPTPEET KAASPVLQED IDIEGVQSEG QDN GAEDSG DTEDELRRVA EQKEHRLPPG QEENGEDPYA GSTDENTDSE EHQEPPDLPV PELP DFFQG KHFFLYGEFP GDERRKLIRY VTAFNGELED NMSDRVQFVI TAQEWDPSFE EALMD NPSL AFVRPRWIYS CNEKQKLLPH QLYGVVPQA |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Details | Co-purified proteins were crosslinked with glutaraldehyde, then buffer exchanged to remove the crosslinker before negative staining. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average exposure time: 1.0 sec. / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 67000 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)