+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22092 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vigna radiata complex I* membrane arm. | |||||||||
 Map data Map data | Vigna radiata complex I* membrane arm. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Vigna radiata (mung bean) Vigna radiata (mung bean) | |||||||||
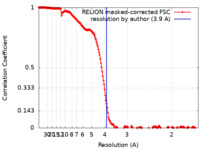
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Letts JA / Maldonado M / Padavannil A / Long Z / Guo F | |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Atomic structure of a mitochondrial complex I intermediate from vascular plants. Authors: Maria Maldonado / Abhilash Padavannil / Long Zhou / Fei Guo / James A Letts /  Abstract: Respiration, an essential metabolic process, provides cells with chemical energy. In eukaryotes, respiration occurs via the mitochondrial electron transport chain (mETC) composed of several large ...Respiration, an essential metabolic process, provides cells with chemical energy. In eukaryotes, respiration occurs via the mitochondrial electron transport chain (mETC) composed of several large membrane-protein complexes. Complex I (CI) is the main entry point for electrons into the mETC. For plants, limited availability of mitochondrial material has curbed detailed biochemical and structural studies of their mETC. Here, we present the cryoEM structure of the known CI assembly intermediate CI* from at 3.9 Å resolution. CI* contains CI's NADH-binding and CoQ-binding modules, the proximal-pumping module and the plant-specific γ-carbonic-anhydrase domain (γCA). Our structure reveals significant differences in core and accessory subunits of the plant complex compared to yeast, mammals and bacteria, as well as the details of the γCA domain subunit composition and membrane anchoring. The structure sheds light on differences in CI assembly across lineages and suggests potential physiological roles for CI* beyond assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22092.map.gz emd_22092.map.gz | 480.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22092-v30.xml emd-22092-v30.xml emd-22092.xml emd-22092.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22092_fsc.xml emd_22092_fsc.xml | 18.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22092.png emd_22092.png | 93.7 KB | ||
| Masks |  emd_22092_msk_1.map emd_22092_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_22092_half_map_1.map.gz emd_22092_half_map_1.map.gz emd_22092_half_map_2.map.gz emd_22092_half_map_2.map.gz | 412.5 MB 412.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22092 http://ftp.pdbj.org/pub/emdb/structures/EMD-22092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22092 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22092 | HTTPS FTP |
-Related structure data
| Related structure data |  6x89C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10586 (Title: Cryo electron micrographs of digitonin-solubilized, amphipol-stabilized, sucrose-gradient-purified V. radiata mitochondrial membranes - mixed fraction containing CI*, CIII2 and SC III2+IV EMPIAR-10586 (Title: Cryo electron micrographs of digitonin-solubilized, amphipol-stabilized, sucrose-gradient-purified V. radiata mitochondrial membranes - mixed fraction containing CI*, CIII2 and SC III2+IVData size: 6.7 TB Data #1: Raw movies of mixed sample used for determination of mung bean respiratory CI*, CIII2, CIV and SC III2+IV [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22092.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22092.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata complex I* membrane arm. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22092_msk_1.map emd_22092_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map for Vigna radiata complex I* membrane arm.
| File | emd_22092_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map for Vigna radiata complex I* membrane arm. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map for Vigna radiata complex I* membrane arm.
| File | emd_22092_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map for Vigna radiata complex I* membrane arm. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vigna radiata complex I* membrane arm
| Entire | Name: Vigna radiata complex I* membrane arm |
|---|---|
| Components |
|
-Supramolecule #1: Vigna radiata complex I* membrane arm
| Supramolecule | Name: Vigna radiata complex I* membrane arm / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Membrane arm of complex I 800 kDa assembly intermediate (complex I*) |
|---|---|
| Source (natural) | Organism:  Vigna radiata (mung bean) Vigna radiata (mung bean) |
-Macromolecule #1: Vigna radiata complex I* membrane arm
| Macromolecule | Name: Vigna radiata complex I* membrane arm / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vigna radiata (mung bean) Vigna radiata (mung bean) |
| Sequence | String: MYIAVPAEI LGIILPLLLG VAFLVLAERK VMAFVQRRKG PDVVGSFGLL QPLADGLKLI L KEPISPSS ANFSLFRMAP VATFMLSLVA RAVVPFDYGM VLSDSNIGLL YLFAISSLGV YG IITAGWS SNSKYAFLGA LRSAAQMVSY EVSIGLILIT VLICVGPRNS ...String: MYIAVPAEI LGIILPLLLG VAFLVLAERK VMAFVQRRKG PDVVGSFGLL QPLADGLKLI L KEPISPSS ANFSLFRMAP VATFMLSLVA RAVVPFDYGM VLSDSNIGLL YLFAISSLGV YG IITAGWS SNSKYAFLGA LRSAAQMVSY EVSIGLILIT VLICVGPRNS SEIVMAQKQI WSG IPLFPV LVMFFISRLA ETNRAPFDLP EAEAESVAGY NVEYSSMGSA LFFLGEYANM ILMS GPCTS LSPGGWPPIL DLPIFKRIPG SIWFSIKVIL FLFLYIWVRA AFPRYRYDQL MGLGR KVFL PLSLARVVAV SGVSVTFPWL P MFNLFLAV YPEIFIINAT SILLIHGVVF STSKKYDYPP LVSNVGWLGL LSVLITLLLL AA GAPLLTI AHLFRNNLFR RDNFTYFCQI LLLLSTAGTI SMCFDSSEQE RFDAFEFIVL IPL PTRSML FMISAHDSIA MYLAIEPQSL CFYVMAASKR KSEFSTEAGS KYLILGAFSS GILL FGCSM IYGSTGATHF DQLAKILTGY EITGARSSGI FMGILSIAVG SLFKITAVPF HMWAP DIYE GSPTPVTAFL SIAPKISIFA NISRVSIYGS YGATLQQIFC FCSIASMILG ALAAMA QTK VKRPLAHSSI GHVGYIRTGF SCGTIEGIQS LLIGIFIYAL MTIDAFAIVL ALRQTRV KY IADLGALAKT NPISAITFSI TMFSYVGIPP LAGFCSKFYL FFAALGCGAY FLAPVGVV T SVIGCFYYIR LAKRMFFDTP RTWILYEPMD RDKSLLLAMT SSFITSSFPY PSPLFSVTH QMALSSYL M SEFAPICIYL VISLLVSLIL LGLPFPFASN SSTYPEKLSA YECGFDPFGD ARSRFDIRF YLVSILFIIP DPEVTFSFPW AVPPNKIDPF GSWSMMAFLL ILTIGSLYEW KRGASDRE M DPIKYFTFSM IISILGIRGI LLNRRNIPIM SMPIESMLLA VNSNFLVFSV SSDDMMGQS FASLVSTVAA AESAIGLAIF VITFRVRGTI AVEFINSIQG MILSVLSSP ALVSGLMVAR AKNPVHSVLF PIPVFRDTSG LLLLLGLDFS AMIFPVVHIG A IAVSFLFV VMMFHIQIAE IHEEVLRYLP VSGIIGLILW WEMFFILDNE TIPLLPTQIN TT SLIYTVY AGKVRSWTNL ETLGNLLYTY YFVWFLVPSL ILLVAMIGAI VLTMHRTTKV KRQ DVFRRN AIDFRRTIMR RTTDPLTID MGTLGRAFYA VGFWIRETGQ AIDRLGSRLQ GNYLFQEQLS RHRPLMNLFD KYPSVHKDAF VAPSASLLG DVHVGPASSI WYGCVLRGDV NGIVIGSGTN IQDNSLVHVA KSNLTGKVLP T IIGDNVTV GHSAVLQGCT VEDEAFIGMG ATLLDGVYVE KHAMVAAGAL VRQNTRIPYG EV WGGNPAK FLRKLTEDEM AFFSQSALNY SNLAQAHAAE NAKELEETEF VKVLHKKFAP RGE EYDSIL DGGQETPAKL NLQDNVLLDK APKA MGTLG RAIYSVGFWI RETGQAIDRL GSRLQGGYFF QEQLSRHRTL MNIFDKAPVV DKDVF VAPS ASVIGDVQVG RGSSIWYGCV LRGDVNSIRV GSGTNIQDNS LVHVAKSNLS GKVLPT VIG DNVTVGHSAV LHGCTVEDEA FVGMGAVLLD GVVVEKNAMV AAGALVRQNT RIPSGEV WA GNPAKFLRKL SNEEITFISQ SAINYTNLAQ VHAAENSKSY DEIEFEKVLR KKYARKDE E YDSMLGVVRE IPPELILPDN VLPDKAEKAL QK MANLARF SKRALRSAHS LVRHAQSQVQ PQLFAEERAF STEAAKSITP SPDRVKWDYR GQR KIIPLG QWLPKVAVDA YVAPNVVLAG QVTVWDGASV WPGCVLRGDL NKISIGFCSN VQER SVLHA AWTSPTGLPA DTSIERYVTI GAYSLLRSCT IEPECIIGQH SILMEGSLVE TQSIL EAGS VVPPGRRIPS GELWAGNPAR FVRTLTHEEI LEIPKLAVAI NDLSRDHYSE FLPYST VYL EVEKFKKSLG ISV MNTDIT ASTKPEYPVV DRNPPFTKVV GNFSTLDYLR FVTITGVSVT VGYLSGIKPG IRGP SMVTG GLIGVMGGFM YAYQNSAGRL MGFFPNDDEV ARHNKK MVL SATTIGALLG LGTQMYSNAL RKLPYMRHPW EHVVGMGLGA VFVNQLLKWE AQVEQDL DK MLEKAKAANE RRYIDGDDD MASAVDASGN PIPSSSVLMA SSKHIGIRCH SENLDFLKCK KKDPNPEKCL DKGRDVTRCV LGLLKDLHQ KCTKEMDDYV GCMYYHTNEF DLCRKEQQAF EKKCSLE MT EAYIRNKPGM VSVKDMPVLQ DGPPPGGFAP VRFARRIPNK GPSAVAIFLA AFGTFSWG M YQVGQGNKIR RALKEEKYAA RRSILPVLQA EEDERFVREW HKYLAYEAEV MKDVPGWKV GESVYNSGRW VPPASGELRP DIW MNLRWM EAVLPLGIIA GMLCVMGNAQ YYIHKAAHGR PKHIGNDLWD VAMERRDKKL HEQA ASSSN MASGWGITG NKGRCYDFWM DFSECMSRCR EPKDCGLLRE DYLECLHHSK EFQRRNRIYK E EQRKIRAA AKKAKDDGVV KEHHYLHGNT TDSNPHLSKV KKMSRRSSRT VYVGNLPGDI RE REVEDLF LKYGHITHID LKVPPRPPGY AFVEFEDAQD AEDAIRGRDG YDFDGHRLRV EPA HGGRGH SSSRDRHSSH SNGRAGRGVS RRSEYRVLVT GLPSSASWQD LKDHMRKAGD VCFS QVFHD GRGTTGIVDY TNYDDMKYAI KKLDDSEFRN AFSKGYVRVR EYDSRRDSSR SPSRG PSHS RGRSYSRSRS RSRSYSRDRS QSKSPKGKSS QRSPPAKSPS RSPAARSRSR SRSRSL SGS RSRSRSPLPP RNKSPKRGSA SRSPSRSRSR SKSLSR MAV MEKLKMFVVQ EPVVAASCLI AGFGLFLPAV VRPILDSYQA TKQPPQPALS DVVAGMT GK K MAARVAAR YGSRRLFSSG SGKILSEEEK AAENAYFKKA EQDKLEKLAR KGPQPEASSG GS VIDAKPS GSGHTGASAE RVSTDKHRNY AVVAGTITIL GALGWYLKGT AKKPEVQD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III Details: Blotted for 9 seconds at 100% humidity, 15 degrees C.. | |||||||||||||||
| Details | Extracted from V. radiata mitochondrial membranes with digitonin, exchanged into amphipole A8-35, purified on sucrose gradient. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9816 / Average exposure time: 3.0 sec. / Average electron dose: 86.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)