+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22091 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vigna radiata mitochondrial complex I* | |||||||||
 Map data Map data | Mitochondrial respiratory complex I* from Vigna radiata. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Vigna radiata (mung bean) Vigna radiata (mung bean) | |||||||||
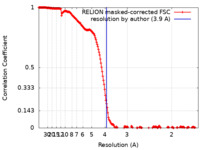
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Letts JA / Maldonado M / Padavannil A / Zhou L / Guo F | |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Atomic structure of a mitochondrial complex I intermediate from vascular plants. Authors: Maria Maldonado / Abhilash Padavannil / Long Zhou / Fei Guo / James A Letts /  Abstract: Respiration, an essential metabolic process, provides cells with chemical energy. In eukaryotes, respiration occurs via the mitochondrial electron transport chain (mETC) composed of several large ...Respiration, an essential metabolic process, provides cells with chemical energy. In eukaryotes, respiration occurs via the mitochondrial electron transport chain (mETC) composed of several large membrane-protein complexes. Complex I (CI) is the main entry point for electrons into the mETC. For plants, limited availability of mitochondrial material has curbed detailed biochemical and structural studies of their mETC. Here, we present the cryoEM structure of the known CI assembly intermediate CI* from at 3.9 Å resolution. CI* contains CI's NADH-binding and CoQ-binding modules, the proximal-pumping module and the plant-specific γ-carbonic-anhydrase domain (γCA). Our structure reveals significant differences in core and accessory subunits of the plant complex compared to yeast, mammals and bacteria, as well as the details of the γCA domain subunit composition and membrane anchoring. The structure sheds light on differences in CI assembly across lineages and suggests potential physiological roles for CI* beyond assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
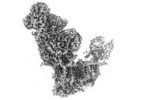
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22091.map.gz emd_22091.map.gz | 480.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22091-v30.xml emd-22091-v30.xml emd-22091.xml emd-22091.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22091_fsc.xml emd_22091_fsc.xml | 18.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22091.png emd_22091.png | 70.1 KB | ||
| Masks |  emd_22091_msk_1.map emd_22091_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_22091_half_map_1.map.gz emd_22091_half_map_1.map.gz emd_22091_half_map_2.map.gz emd_22091_half_map_2.map.gz | 412.1 MB 412.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22091 http://ftp.pdbj.org/pub/emdb/structures/EMD-22091 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22091 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22091 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22091.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22091.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mitochondrial respiratory complex I* from Vigna radiata. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
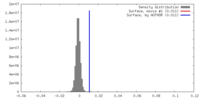
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22091_msk_1.map emd_22091_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
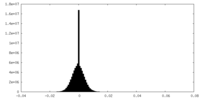
| Density Histograms |
-Half map: Vigna radiata mitochondrial complex
| File | emd_22091_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata mitochondrial complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
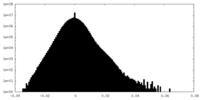
| Density Histograms |
-Half map: Vigna radiata mitochondrial complex
| File | emd_22091_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Vigna radiata mitochondrial complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vigna radiata complex I*
| Entire | Name: Vigna radiata complex I* |
|---|---|
| Components |
|
-Supramolecule #1: Vigna radiata complex I*
| Supramolecule | Name: Vigna radiata complex I* / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Complex I 800 kDa assembly intermediate (complex I*) |
|---|---|
| Source (natural) | Organism:  Vigna radiata (mung bean) Vigna radiata (mung bean) |
| Molecular weight | Theoretical: 800 KDa |
-Macromolecule #1: Vigna radiata complex I*
| Macromolecule | Name: Vigna radiata complex I* / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vigna radiata (mung bean) Vigna radiata (mung bean) |
| Sequence | String: MGFGLLASK ALRPTSRLLL SSQNPTIFLR TIVSKPQLCN PEASAAQPAQ PPPVDLPPRS P LAGARVHF ANPDDAIEVF VDGYPVKIPK GMTVLQACEV AGVDIPRFCY HSRLSIAGNC RM CLVEVEK SPKPVASCAM PALPGMKIKT DTPVAKKARE GVMEFLLMNH ...String: MGFGLLASK ALRPTSRLLL SSQNPTIFLR TIVSKPQLCN PEASAAQPAQ PPPVDLPPRS P LAGARVHF ANPDDAIEVF VDGYPVKIPK GMTVLQACEV AGVDIPRFCY HSRLSIAGNC RM CLVEVEK SPKPVASCAM PALPGMKIKT DTPVAKKARE GVMEFLLMNH PLDCPICDQG GEC DLQDQS MAFGSDRGRF TEVKRSVVDK NLGPLVKTVM TRCIQCTRCV RFATEVAGVQ DLGM LGRGS GEEIGTYVEK LLTSELSGNV IDICPVGALT SKPFAFKARN WELKGTETID VTDAV GSNI RIDSRGPEVM RIVPRLNEDI NEEWISDKTR FCYDGLKRQR LNDPMIRGPD GRFKAV NWR DALSVIADIA HQVKPEEIVG VAGKLSDAES MIALKDFLNR MGSNDVWGEG IGVNTNA DF RSGYIMNTSI AGLEKADVFL LVGTQPRVEA AMVNARIRKT VRSNQAKVGY IGPATDFN Y DHKHLGTDPQ TLVEIAEGRH PFFKTLSDAK NPVIIVGAGV FERKDQDAIF AAVETIAQK ANVVRPDWNG LNVLLLHAAQ AAALDLGLVP QSEKSLESAK FVYLMGADDV NLDKIPDDAF VVYQGHHGD KSVYRANVIL PTAAFSEKEG TYQNTEGCTQ QTLPAVPTVG DSRDDWKIIR A LSEVAGVR LPYDTIGAVR ARIRNVAPNL VNVDEREPAT LPSSLRPSFT QKVDTTPFGT VI ENFYMTD AITRASKIMA QCSATLLKK MTTRNGQIKN FTSNFGPQHP AAHGVSRSVL EMNGEVVERA EPHIGSLHRG TEKLIEYKTY LQALPYSDR LDYVSTMAQE HAHSSAVERL LNCEVPLRAQ YIRVLFREIT RISNHLLALT T HAMDVGAS TPSLWAFEER EKLLEFYERV SGARMHASFI RPGGVAQDLP LGLCRDIDSF TQ QFASRID ELEEMSTGNR IWKQRLVDIG TVTAQQAKDW GFSGVMLRGP GVCWDSRRAA PYD VHDQSD PDVPVGTRGD RYDRYCIRIE EMRQSMRIIW QCPNKIPSGM IKADDRKLCP PSRC RMKLS MESSIHHFEL YTEGFSVPAP STYTAVEAPK GEFGVFLVSN GSNRPYRRKI RAPGS AHSQ GLDSMSKHHM PADVVTIIGT QDIVSGEVDR MDNQSIFKY SWETLPKKWV KKMERSEHGN RSDTKTDYLF QLLCFLKLHT YTRVQVSIDI C GVDYPSRK RRFEVVYNLL STRYNSRIRV QTSADEVTRI SPVVSLFPSA GRWEREVWDM FG VSSINHP DLRRISTDYG FEGHPLRKDL PLSGYVEVRY DDPEKRVVSE PIEMTQEFRY FDF ASPWEQ RSDG MALLS RASSRIYQVH SYQRALSLHT TLPALNASTS THTPTPYAPP PPPSASSPVG VSKAA EFVI SKVDDLMNWA RRGSIWPMTF GLACCAVEMM HTGAARYDLD RFGIIFRPSP RQSDCM IVA GTLTNKMAPA LRKVYDQMPE PRWVISMGSC ANGGGYYHYS YSVVRGCDRI VPVDIYV PG CPPTAEALLY GLLQLQKKIN RRKDFLHWWT K MAAILARK SLLALRARQL AVSGQGLHSS QSYGLRLSAH SFSTKLEDEQ REQLAKEISK DW SSVFERS INTLFLTEMV RGLMLTLKYF FETKVTINYP FEKGPLSPRF RGEHALRRYP TGE ERCIAC KLCEAICPAQ AITIEAEERE DGSRRTTRYD IDMTKCIYCG FCQEACPVDA IVEG PNFEF ATETHEELLY DKEKLLENGD RWETEIAENL RSESLYR MR GILSLRSALV QHRSEKLGLG LRQFSTQGAS TPTSPQPPPP PASPQPPPPP PPPEKTHF G GLKDEDRIFT NLYGLHDPFL KGAMKRGDWY RTKDLVIKGT DWIVNEMKKS GLRGRGGAG FPSGLKWSFM PKVSDGRPSY LVVNADESEP GTCKDREIMR HDPHKLLEGC LIAGVGMRAT AAYIYIRGE YVNERKNLEK ARQEAYAAGL LGKNACGSGY DFDVHIHFGA GAYICGEETA L LESLEGKQ GKPRLKPPFP ANAGLYGCPT TVTNVETVAV SPTILRRGPE WFAGFGRKNN AG TKLFCVS GHVNKPCTVE EEMSIPLKEL IERHCGGVRG GWDNLLAVIP GGSSVPLLPK SIC DDVLMD YDALKAVQSG LGTAAVIVMD KSTDVVDAIA RLSYFYKHES CGQCTPCREG TGWL WTIME RMKVGNAKLE EIDMLQELTK QIEGHTICAL GDAAAWPVQG LIRHFRPELE RRIRE NAER ELLQATG ML ARLAANRFNE IRHIFRQPSR AFSTALNYHL DSPDNNPNLP WEFTEPNKAK VTEILSHY P SNYKQSAVIP LLDLAQQQHG GWLPVSAMNA VANIIEVPPI RVYEVATFYS MFNRAKVGK YHLLVCGTTP CMIRGSRGIE EALLKHLGVK RNEVTPDGLF SVGEMECMGC CVNAPMITVA DYSNGSEGY TYNYYEDVTP EKVVEIVEKL RKGEKPPHGT QNPLRIRSGP EGGNTTLLGE P KPPPCRDL DAC MAWRGQ LSKNIKELRF LMCQSSSASA SARAFVEKNY KELKTLNPKL PILIRECSGV EPQL WARYD LGVEKAIKLE GLSEAQISKA LEDLAKAGQS SKA MFLRAV ARPLMAKVKA STGIVGLEVV PNAREVLIGL YSKTLKEIQK VPEDEGYRKA VESF TKHRL SVCKEEEDWE AIEKRLGCGQ VEELIEEAQD ELKLISYMIE WDPWGVPDDY ECEVI ENDA PVPKHVPLHR PPPLPTEFHK TLEALSSQSG KDIPGASSTE SPSKT MASA LRNVKVLPNS ASVEEARQRV FDFFRATCRS LPSVMEIYNL YDVTSVAQLR STVASE IRK NIGVSDPKVI DMLLFKGMEE LMNVVNHSKQ RHHIIGQYVV GQQGQVQDSA SKDPAIS PF LKNFYSSNYF MAKSASNSL VQTLKRYIKK PWEITGPCAD PEYRSAVPLA TEYRLQCPAT TKEKPCIPNS L PETVYDIK YFSRDQRRNR PPIRRTVLKK ADVEKLAKEQ TFAVSDFPPV YLNSAVEEDI NA IGGGYQG MQTIARRLG HQSLKSSTSI RAIYPISDHH YVADHQRYVS TIATKGVGHL VRKGTGGRSS V SGIIATVF GATGFLGRYV VQQLAKMGSQ VLVPFRGSED CPRHLKLMGD LGQIVPMKYN PR DESSVKA VMAKANVVIN LIGRDYETRN YSFEEVHYHM AENLAKISRE HGGILRFIQV SCL GASPSS PSRMLRAKAA AEEVVLRELP EATILKPAVM IGTEDRILNP WAHFAKKYGF IPLF GNGST KIQPVYVVDV AAALTTVLKD DGTSMGKTYE LGGPEIFTVH DLAELMYETI REWPR YVKV PFPIAKALAT PREILLNKVP FPLPTPEILN LDKIQALTTD TIVSENALTF NDLGII PHK LKGYPVEFLI SYRKGGPQFG STISERVTPD AWP MASVVK NVLKSIREKG FGTFLRELKD EGYLRCLPDG NLLQTKIHNI GATLVGVDKF GNKY YEKLG DTQYGRHRWV EYAEKSRYNA SQVPPEWHGW LHFITDHTGD ELLLLKPKRY GVEHK ENLS GEGEQYIYHS KGHALNPGQR DWTRYQPWES KA MANTLQR ALRATMARRF STQALVETKP GEIGMVSGIP QEHLRRRVVI YSPARTASQQ GSG KVGRWK INFLSTQKWE NPLMGWTSTG DPYAHVGDSA LTFDSAEAAK AFAEKHGWEY SVKK RHTPL LKVKSYADNF KWKGFPKPAE E MASSVLRN LMRFSSPRNT STRSFSLVTS QISNHTAKWM QDTSKKSPME LINEVPPIKV EG RIVACEG DTNPALGHPI EFICLDLPEP AVCKYCGLRY VQDHHH MYI AVPAEILGII LPLLLGVAFL VLAERKVMAF VQRRKGPDVV GSFGLLQPLA DGLKLIL KE PISPSSANFS LFRMAPVATF MLSLVARAVV PFDYGMVLSD SNIGLLYLFA ISSLGVYG I ITAGWSSNSK YAFLGALRSA AQMVSYEVSI GLILITVLIC VGPRNSSEIV MAQKQIWSG IPLFPVLVMF FISRLAETNR APFDLPEAEA ESVAGYNVEY SSMGSALFFL GEYANMILMS GPCTSLSPG GWPPILDLPI FKRIPGSIWF SIKVILFLFL YIWVRAAFPR YRYDQLMGLG R KVFLPLSL ARVVAVSGVS VTFPWLP MY IAVPAEILGI ILPLLLGVAF LVLAERKVMA FVQRRKGPDV VGSFGLLQPL ADGLKLIL K EPISPSSANF SLFRMAPVAT FMLSLVARAV VPFDYGMVLS DSNIGLLYLF AISSLGVYG IITAGWSSNS KYAFLGALRS AAQMVSYEVS IGLILITVLI CVGPRNSSEI VMAQKQIWSG IPLFPVLVM FFISRLAETN RAPFDLPEAE AESVAGYNVE YSSMGSALFF LGEYANMILM S GPCTSLSP GGWPPILDLP IFKRIPGSIW FSIKVILFLF LYIWVRAAFP RYRYDQLMGL GR KVFLPLS LARVVAVSGV SVTFPWLP M FNLFLAVYPE IFIINATSIL LIHGVVFSTS KKYDYPPLVS NVGWLGLLSV LITLLLLAA GAPLLTIAHL FRNNLFRRDN FTYFCQILLL LSTAGTISMC FDSSEQERFD AFEFIVLIPL PTRSMLFMI SAHDSIAMYL AIEPQSLCFY VMAASKRKSE FSTEAGSKYL ILGAFSSGIL L FGCSMIYG STGATHFDQL AKILTGYEIT GARSSGIFMG ILSIAVGSLF KITAVPFHMW AP DIYEGSP TPVTAFLSIA PKISIFANIS RVSIYGSYGA TLQQIFCFCS IASMILGALA AMA QTKVKR PLAHSSIGHV GYIRTGFSCG TIEGIQSLLI GIFIYALMTI DAFAIVLALR QTRV KYIAD LGALAKTNPI SAITFSITMF SYVGIPPLAG FCSKFYLFFA ALGCGAYFLA PVGVV TSVI GCFYYIRLAK RMFFDTPRTW ILYEPMDRDK SLLLAMTSSF ITSSFPYPSP LFSVTH QMA LSSYL MSEF APICIYLVIS LLVSLILLGL PFPFASNSST YPEKLSAYEC GFDPFGDARS RFDIRF YLV SILFIIPDPE VTFSFPWAVP PNKIDPFGSW SMMAFLLILT IGSLYEWKRG ASDRE MDPI KYFTFSMIIS ILGIRGILLN RRNIPIMSMP IESMLLAVNS NFLVFSVSSD DMMGQS FAS LVSTVAAAES AIGLAIFVIT FRVRGTIAVE FINSIQG MI LSVLSSPALV SGLMVARAKN PVHSVLFPIP VFRDTSGLLL LLGLDFSAMI FPVVHIGA I AVSFLFVVMM FHIQIAEIHE EVLRYLPVSG IIGLILWWEM FFILDNETIP LLPTQINTT SLIYTVYAGK VRSWTNLETL GNLLYTYYFV WFLVPSLILL VAMIGAIVLT MHRTTKVKRQ DVFRRNAID FRRTIMRRTT DPLTID MGT LGRAFYAVGF WIRETGQAID RLGSRLQGNY LFQEQLSRHR PLMNLFDKYP SVHKDAF VA PSASLLGDVH VGPASSIWYG CVLRGDVNGI VIGSGTNIQD NSLVHVAKSN LTGKVLPT I IGDNVTVGHS AVLQGCTVED EAFIGMGATL LDGVYVEKHA MVAAGALVRQ NTRIPYGEV WGGNPAKFLR KLTEDEMAFF SQSALNYSNL AQAHAAENAK ELEETEFVKV LHKKFAPRGE EYDSILDGG QETPAKLNLQ DNVLLDKAPK A MGTLGRAI YSVGFWIRET GQAIDRLGSR LQGGYFFQEQ LSRHRTLMNI FDKAPVVDKD VF VAPSASV IGDVQVGRGS SIWYGCVLRG DVNSIRVGSG TNIQDNSLVH VAKSNLSGKV LPT VIGDNV TVGHSAVLHG CTVEDEAFVG MGAVLLDGVV VEKNAMVAAG ALVRQNTRIP SGEV WAGNP AKFLRKLSNE EITFISQSAI NYTNLAQVHA AENSKSYDEI EFEKVLRKKY ARKDE EYDS MLGVVREIPP ELILPDNVLP DKAEKALQK MANLARFSKR ALRSAHSLVR HAQSQVQPQL FAEERAFSTE AAKSITPSPD RVKWDYRGQR KIIPLGQWL PKVAVDAYVA PNVVLAGQVT VWDGASVWPG CVLRGDLNKI SIGFCSNVQE R SVLHAAWT SPTGLPADTS IERYVTIGAY SLLRSCTIEP ECIIGQHSIL MEGSLVETQS IL EAGSVVP PGRRIPSGEL WAGNPARFVR TLTHEEILEI PKLAVAINDL SRDHYSEFLP YST VYLEVE KFKKSLGISV MNTDITAST KPEYPVVDRN PPFTKVVGNF STLDYLRFVT ITGVSVTVGY LSGIKPGIRG P SMVTGGLI GVMGGFMYAY QNSAGRLMGF FPNDDEVARH NKK MVLSAT TIGALLGLGT QMYSNALRKL PYMRHPWEHV VGMGLGAVFV NQLLKWEAQV EQDL DKMLE KAKAANERRY IDGDDD MAS AVDASGNPIP SSSVLMASSK HIGIRCHSEN LDFLKCKKKD PNPEKCLDKG RDVTRCV LG LLKDLHQKCT KEMDDYVGCM YYHTNEFDLC RKEQQAFEKK CSLE MTEAY IRNKPGMVSV KDMPVLQDGP PPGGFAPVRF ARRIPNKGPS AVAIFLAAFG TFSWG MYQV GQGNKIRRAL KEEKYAARRS ILPVLQAEED ERFVREWHKY LAYEAEVMKD VPGWKV GES VYNSGRWVPP ASGELRPDIW MNLRWMEAV LPLGIIAGML CVMGNAQYYI HKAAHGRPKH IGNDLWDVAM ERRDKKLHEQ A ASSSN MA SGWGITGNKG RCYDFWMDFS ECMSRCREPK DCGLLREDYL ECLHHSKEFQ RRNRIYKE E QRKIRAAAKK AKDDGVVKEH HYLHGNTTDS NPHLSKVKKM SRRSSRTVYV GNLPGDIRE REVEDLFLKY GHITHIDLKV PPRPPGYAFV EFEDAQDAED AIRGRDGYDF DGHRLRVEPA HGGRGHSSS RDRHSSHSNG RAGRGVSRRS EYRVLVTGLP SSASWQDLKD HMRKAGDVCF S QVFHDGRG TTGIVDYTNY DDMKYAIKKL DDSEFRNAFS KGYVRVREYD SRRDSSRSPS RG PSHSRGR SYSRSRSRSR SYSRDRSQSK SPKGKSSQRS PPAKSPSRSP AARSRSRSRS RSL SGSRSR SRSPLPPRNK SPKRGSASRS PSRSRSRSKS LSR MAVMEK LKMFVVQEPV VAASCLIAGF GLFLPAVVRP ILDSYQATKQ PPQPALSDVV AGMT GKK M AARVAARYGS RRLFSSGSGK ILSEEEKAAE NAYFKKAEQD KLEKLARKGP QPEASSGGS VIDAKPSGSG HTGASAERVS TDKHRNYAVV AGTITILGAL GWYLKGTAKK PEVQD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Details: 30 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III Details: Blotted for 9 seconds at 100% humidity, 15 degrees C.. | |||||||||||||||
| Details | Extracted from V. radiata mitochondrial membranes with digitonin, exchanged into amphipole A8-35, purified on sucrose gradient. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 9816 / #0 - Average exposure time: 3.0 sec. / #0 - Average electron dose: 86.4 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Number grids imaged: 1 / #1 - Number real images: 9816 / #1 - Average exposure time: 3.0 sec. / #1 - Average electron dose: 86.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)