+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21910 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The yeast Ctf3 complex with Cnn1-Wip1 | |||||||||
 Map data Map data | CMMCW | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinetochore / cell cycle / chromosome segregation / CCAN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of kinetochore assembly / attachment of spindle microtubules to kinetochore / centromeric DNA binding / establishment of mitotic sister chromatid cohesion / mitotic spindle assembly checkpoint signaling / DNA replication initiation / meiotic cell cycle / chromosome segregation / kinetochore / cell division ...negative regulation of kinetochore assembly / attachment of spindle microtubules to kinetochore / centromeric DNA binding / establishment of mitotic sister chromatid cohesion / mitotic spindle assembly checkpoint signaling / DNA replication initiation / meiotic cell cycle / chromosome segregation / kinetochore / cell division / protein-containing complex binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
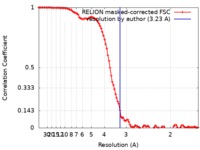
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Hinshaw SM / Harrison SC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Curr Biol / Year: 2020 Journal: Curr Biol / Year: 2020Title: The Structural Basis for Kinetochore Stabilization by Cnn1/CENP-T. Authors: Stephen M Hinshaw / Stephen C Harrison /  Abstract: Chromosome segregation depends on a regulated connection between spindle microtubules and centromeric DNA. The kinetochore mediates this connection and ensures it persists during anaphase, when ...Chromosome segregation depends on a regulated connection between spindle microtubules and centromeric DNA. The kinetochore mediates this connection and ensures it persists during anaphase, when sister chromatids must transit into daughter cells uninterrupted. The Ctf19 complex (Ctf19c) forms the centromeric base of the kinetochore in budding yeast. Biochemical experiments show that Ctf19c members associate hierarchically when purified from cell extract [1], an observation that is mostly explained by the structure of the complex [2]. The Ctf3 complex (Ctf3c), which is not required for the assembly of most other Ctf19c factors, disobeys the biochemical assembly hierarchy when observed in dividing cells that lack more basal components [3]. Thus, the biochemical experiments do not completely recapitulate the logic of centromeric Ctf19c assembly. We now present a high-resolution structure of the Ctf3c bound to the Cnn1-Wip1 heterodimer. Associated live-cell imaging experiments provide a mechanism for Ctf3c and Cnn1-Wip1 recruitment to the kinetochore. The mechanism suggests feedback regulation of Ctf19c assembly and unanticipated similarities in kinetochore organization between yeast and vertebrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21910.map.gz emd_21910.map.gz | 77.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21910-v30.xml emd-21910-v30.xml emd-21910.xml emd-21910.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21910_fsc.xml emd_21910_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_21910.png emd_21910.png | 172.4 KB | ||
| Filedesc metadata |  emd-21910.cif.gz emd-21910.cif.gz | 6.9 KB | ||
| Others |  emd_21910_additional.map.gz emd_21910_additional.map.gz | 77.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21910 http://ftp.pdbj.org/pub/emdb/structures/EMD-21910 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21910 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21910 | HTTPS FTP |
-Related structure data
| Related structure data |  6wucMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21910.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21910.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CMMCW | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: CMMCW-N
| File | emd_21910_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CMMCW-N | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Single particle reconstruction of the Ctf3c bound to Cnn1-Wip1
| Entire | Name: Single particle reconstruction of the Ctf3c bound to Cnn1-Wip1 |
|---|---|
| Components |
|
-Supramolecule #1: Single particle reconstruction of the Ctf3c bound to Cnn1-Wip1
| Supramolecule | Name: Single particle reconstruction of the Ctf3c bound to Cnn1-Wip1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Inner kinetochore subunit MCM16
| Macromolecule | Name: Inner kinetochore subunit MCM16 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.438359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMTNSSEK QWERIQQLEK EHVEVYRELL ITLDRLYLIR KHNHAVILSH TQQRLLEIRH QLQINLEKTA LLIRLLEKPD NTNVLFTKL QNLLEESNSL DYELLQSLGA QSSLHKQLIE SRAERDELMS KLIELSSKFP KPTIPPDDSD TAGKQVEVEK E NETIQELM IALQIHSGYT NISYTI UniProtKB: Inner kinetochore subunit MCM16 |
-Macromolecule #2: Inner kinetochore subunit CTF3
| Macromolecule | Name: Inner kinetochore subunit CTF3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.617891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMSLILDD IILSLTNANE RTPPQALKTT LSLLYEKSKQ YGLSSPQLQA LVRLLCETSI IDTVTKVYIV ENCFLPDGYL TKELLLEII NHLGTPTVFS RYRIQTPPVL QSALCKWLVH VYFLFPVHSE REHNISSSIW LHLWQFSFLQ KWITPLVIWQ A TTPVDVKP ...String: SNAMSLILDD IILSLTNANE RTPPQALKTT LSLLYEKSKQ YGLSSPQLQA LVRLLCETSI IDTVTKVYIV ENCFLPDGYL TKELLLEII NHLGTPTVFS RYRIQTPPVL QSALCKWLVH VYFLFPVHSE REHNISSSIW LHLWQFSFLQ KWITPLVIWQ A TTPVDVKP WKLSIIKRCA MHPGYRDAPG SATLILQRFQ CLVGASSQIT ESIITINCNR KTLKSHRNLK LDAHFLSILK RI LSRAHPA NFPADTVQNT IDMYLSEIHQ LGADSIYPLR LQSLPEYVPS DSTVSLWDVT SLEQLAQNWP QLHIPNDVDY MMK PSLNSN VLLPRKVMSR DSLKHLYSSI ILIKNSRDES SSPYEWCIWQ LKRCFAHQIE TPQEVIPIII SVSSMDNKLS SRII QTFCN LKYLKLDELT LKKVCGGILP LWKPELISGT REFFVKFMAS IFMWSTRDGH DNNCTFSETC FYVLQMITNW VLDDK LIAL GLTLLHDMQS LLTLDKIFNN ATSNRFSTMA FISSLDILTQ LSKQTKSDYA IQYLIVGPDI MNKVFSSDDP LLLSAA CRY LVATKNKLMQ YPSTNKFVRM QNQYIMDLTN YLYRNKVLSS KSLFGVSPDF FKQILENLYI PTADFKNAKF FTITGIP AL SYICIIILRR LETAENTKIK FTSGIINEET FNNFFRVHHD EIGQHGWIKG VNNIHDLRVK ILMHLSNTAN PYRDIAAF L FTYLKSLSKY SVQNS UniProtKB: Inner kinetochore subunit CTF3 |
-Macromolecule #3: Inner kinetochore subunit MCM22
| Macromolecule | Name: Inner kinetochore subunit MCM22 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.874799 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMDVEKDV LDVYIKNLEN QIGNKRYFLK QAQGAIDEIT KRSLDTEGKP VNSEVFTELL RKPMFFSERA DPIGFSLTSN FLSLRAQSS SEWLSLMNDQ SVDQKAMLLL QNNINSDLKE LLRKLQHQMT IMDSKKQDHA HIRTRKARNK ELWDSLADFL K GYLVPNLD ...String: SNAMDVEKDV LDVYIKNLEN QIGNKRYFLK QAQGAIDEIT KRSLDTEGKP VNSEVFTELL RKPMFFSERA DPIGFSLTSN FLSLRAQSS SEWLSLMNDQ SVDQKAMLLL QNNINSDLKE LLRKLQHQMT IMDSKKQDHA HIRTRKARNK ELWDSLADFL K GYLVPNLD DNDESIDSLT NEVMLLMKRL IEHDLNLTLN DFSSKTIPIY RLLLRANIIT VIEGSTNPGT KYIKLIDFNE TS LT UniProtKB: Inner kinetochore subunit MCM22 |
-Macromolecule #4: Inner kinetochore subunit WIP1
| Macromolecule | Name: Inner kinetochore subunit WIP1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.527717 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: SNAMDTEALA NYLLRQLSLD AEENKLEDLL QRQNEDQESS QEYNKKLLLA CGFQAILRKI LLDARTRATA EGLREVYPYH IEAATQAFL DSQ UniProtKB: Inner kinetochore subunit WIP1 |
-Macromolecule #5: Inner kinetochore subunit CNN1
| Macromolecule | Name: Inner kinetochore subunit CNN1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.359785 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSTPRKAAGN NENTEVSEIR TPFRERALEE QRLKDEVLIR NTPGYRKLLS ASTKSHDILN KDPNEVRSFL QDLSQVLARK SQGNDTTTN KTQARNLIDE LAYEESQPEE NELLRSRSEK LTDNNIGNET QPDYTSLSQT VFAKLQERDK GLKSRKIDPI I IQDVPTTG ...String: MSTPRKAAGN NENTEVSEIR TPFRERALEE QRLKDEVLIR NTPGYRKLLS ASTKSHDILN KDPNEVRSFL QDLSQVLARK SQGNDTTTN KTQARNLIDE LAYEESQPEE NELLRSRSEK LTDNNIGNET QPDYTSLSQT VFAKLQERDK GLKSRKIDPI I IQDVPTTG HEDELTVHSP DKANSISMEV LRTSPSIGMD QVDEPPVRDP VPISITQQEE PLSEDLPSDD KEETEEAENE DY SFENTSD ENLDDIGNDP IRLNVPAVRR SSIKPLQIMD LKHLTRQFLN ENRIILPKQT WSTIQEESLN IMDFLKQKIG TLQ KQELVD SFIDMGIINN VDDMFELAHE LLPLELQSRI ESYLF UniProtKB: Inner kinetochore subunit CNN1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)