+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20200 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the yeast Ctf3 complex | |||||||||
 Map data Map data | Final volume | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | kinetochore / chromosome / cryo-em / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationattachment of spindle microtubules to kinetochore / establishment of mitotic sister chromatid cohesion / mitotic spindle assembly checkpoint signaling / DNA replication initiation / meiotic cell cycle / chromosome segregation / kinetochore / cell division / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
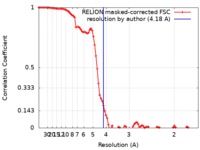
| Method | single particle reconstruction / cryo EM / Resolution: 4.18 Å | |||||||||
 Authors Authors | Hinshaw SM / Harrison SC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: The structure of the yeast Ctf3 complex. Authors: Stephen M Hinshaw / Andrew N Dates / Stephen C Harrison /  Abstract: Kinetochores are the chromosomal attachment points for spindle microtubules. They are also signaling hubs that control major cell cycle transitions and coordinate chromosome folding. Most well- ...Kinetochores are the chromosomal attachment points for spindle microtubules. They are also signaling hubs that control major cell cycle transitions and coordinate chromosome folding. Most well-studied eukaryotes rely on a conserved set of factors, which are divided among two loosely-defined groups, for these functions. Outer kinetochore proteins contact microtubules or regulate this contact directly. Inner kinetochore proteins designate the kinetochore assembly site by recognizing a specialized nucleosome containing the H3 variant Cse4/CENP-A. We previously determined the structure, resolved by cryo-electron microscopy (cryo-EM), of the yeast Ctf19 complex (Ctf19c, homologous to the vertebrate CCAN), providing a high-resolution view of inner kinetochore architecture (Hinshaw and Harrison, 2019). We now extend these observations by reporting a near-atomic model of the Ctf3 complex, the outermost Ctf19c sub-assembly seen in our original cryo-EM density. The model is sufficiently well-determined by the new data to enable molecular interpretation of Ctf3 recruitment and function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20200.map.gz emd_20200.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20200-v30.xml emd-20200-v30.xml emd-20200.xml emd-20200.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20200_fsc.xml emd_20200_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20200.png emd_20200.png | 102.7 KB | ||
| Masks |  emd_20200_msk_1.map emd_20200_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20200.cif.gz emd-20200.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20200 http://ftp.pdbj.org/pub/emdb/structures/EMD-20200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20200 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20200 | HTTPS FTP |
-Validation report
| Summary document |  emd_20200_validation.pdf.gz emd_20200_validation.pdf.gz | 554.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20200_full_validation.pdf.gz emd_20200_full_validation.pdf.gz | 554.1 KB | Display | |
| Data in XML |  emd_20200_validation.xml.gz emd_20200_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_20200_validation.cif.gz emd_20200_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20200 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20200 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20200 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20200 | HTTPS FTP |
-Related structure data
| Related structure data |  6ouaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20200.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20200.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final volume | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_20200_msk_1.map emd_20200_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ctf3 complex
| Entire | Name: Ctf3 complex |
|---|---|
| Components |
|
-Supramolecule #1: Ctf3 complex
| Supramolecule | Name: Ctf3 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Inner kinetochore subunit CTF3
| Macromolecule | Name: Inner kinetochore subunit CTF3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 84.617891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMSLILDD IILSLTNANE RTPPQALKTT LSLLYEKSKQ YGLSSPQLQA LVRLLCETSI IDTVTKVYIV ENCFLPDGYL TKELLLEII NHLGTPTVFS RYRIQTPPVL QSALCKWLVH VYFLFPVHSE REHNISSSIW LHLWQFSFLQ KWITPLVIWQ A TTPVDVKP ...String: SNAMSLILDD IILSLTNANE RTPPQALKTT LSLLYEKSKQ YGLSSPQLQA LVRLLCETSI IDTVTKVYIV ENCFLPDGYL TKELLLEII NHLGTPTVFS RYRIQTPPVL QSALCKWLVH VYFLFPVHSE REHNISSSIW LHLWQFSFLQ KWITPLVIWQ A TTPVDVKP WKLSIIKRCA MHPGYRDAPG SATLILQRFQ CLVGASSQIT ESIITINCNR KTLKSHRNLK LDAHFLSILK RI LSRAHPA NFPADTVQNT IDMYLSEIHQ LGADSIYPLR LQSLPEYVPS DSTVSLWDVT SLEQLAQNWP QLHIPNDVDY MMK PSLNSN VLLPRKVMSR DSLKHLYSSI ILIKNSRDES SSPYEWCIWQ LKRCFAHQIE TPQEVIPIII SVSSMDNKLS SRII QTFCN LKYLKLDELT LKKVCGGILP LWKPELISGT REFFVKFMAS IFMWSTRDGH DNNCTFSETC FYVLQMITNW VLDDK LIAL GLTLLHDMQS LLTLDKIFNN ATSNRFSTMA FISSLDILTQ LSKQTKSDYA IQYLIVGPDI MNKVFSSDDP LLLSAA CRY LVATKNKLMQ YPSTNKFVRM QNQYIMDLTN YLYRNKVLSS KSLFGVSPDF FKQILENLYI PTADFKNAKF FTITGIP AL SYICIIILRR LETAENTKIK FTSGIINEET FNNFFRVHHD EIGQHGWIKG VNNIHDLRVK ILMHLSNTAN PYRDIAAF L FTYLKSLSKY SVQNS UniProtKB: Inner kinetochore subunit CTF3 |
-Macromolecule #2: Inner kinetochore subunit MCM16
| Macromolecule | Name: Inner kinetochore subunit MCM16 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.438359 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMTNSSEK QWERIQQLEK EHVEVYRELL ITLDRLYLIR KHNHAVILSH TQQRLLEIRH QLQINLEKTA LLIRLLEKPD NTNVLFTKL QNLLEESNSL DYELLQSLGA QSSLHKQLIE SRAERDELMS KLIELSSKFP KPTIPPDDSD TAGKQVEVEK E NETIQELM IALQIHSGYT NISYTI UniProtKB: Inner kinetochore subunit MCM16 |
-Macromolecule #3: Inner kinetochore subunit MCM22
| Macromolecule | Name: Inner kinetochore subunit MCM22 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.874799 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMDVEKDV LDVYIKNLEN QIGNKRYFLK QAQGAIDEIT KRSLDTEGKP VNSEVFTELL RKPMFFSERA DPIGFSLTSN FLSLRAQSS SEWLSLMNDQ SVDQKAMLLL QNNINSDLKE LLRKLQHQMT IMDSKKQDHA HIRTRKARNK ELWDSLADFL K GYLVPNLD ...String: SNAMDVEKDV LDVYIKNLEN QIGNKRYFLK QAQGAIDEIT KRSLDTEGKP VNSEVFTELL RKPMFFSERA DPIGFSLTSN FLSLRAQSS SEWLSLMNDQ SVDQKAMLLL QNNINSDLKE LLRKLQHQMT IMDSKKQDHA HIRTRKARNK ELWDSLADFL K GYLVPNLD DNDESIDSLT NEVMLLMKRL IEHDLNLTLN DFSSKTIPIY RLLLRANIIT VIEGSTNPGT KYIKLIDFNE TS LT UniProtKB: Inner kinetochore subunit MCM22 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Grid | Model: C-flat-2/2 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
| Details | Recombinant protein frozen in vitreous ice |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 50.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6oua: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)