[English] 日本語
 Yorodumi
Yorodumi- EMDB-21663: Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21663 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protein-DNA complex / ATPase / DNA-binding protein / Genome organization / Sister chromatid cohesion / Transcription regulation / CELL CYCLE / CELL CYCLE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationeye morphogenesis / external genitalia morphogenesis / gallbladder development / SMC loading complex / Scc2-Scc4 cohesin loading complex / ear morphogenesis / regulation of hair cycle / cohesin loader activity / response to DNA damage checkpoint signaling / maintenance of mitotic sister chromatid cohesion ...eye morphogenesis / external genitalia morphogenesis / gallbladder development / SMC loading complex / Scc2-Scc4 cohesin loading complex / ear morphogenesis / regulation of hair cycle / cohesin loader activity / response to DNA damage checkpoint signaling / maintenance of mitotic sister chromatid cohesion / forelimb morphogenesis / embryonic viscerocranium morphogenesis / negative regulation of mitotic metaphase/anaphase transition / Cohesin Loading onto Chromatin / meiotic cohesin complex / Establishment of Sister Chromatid Cohesion / establishment of meiotic sister chromatid cohesion / uterus morphogenesis / cohesin complex / mitotic cohesin complex / positive regulation of sister chromatid cohesion / regulation of developmental growth / establishment of protein localization to chromatin / negative regulation of glial cell apoptotic process / embryonic digestive tract morphogenesis / chromo shadow domain binding / negative regulation of G2/M transition of mitotic cell cycle / positive regulation of neuron migration / mediator complex binding / establishment of mitotic sister chromatid cohesion / replication-born double-strand break repair via sister chromatid exchange / integrator complex / cellular response to X-ray / metanephros development / lateral element / positive regulation of multicellular organism growth / chromatin looping / positive regulation of ossification / embryonic forelimb morphogenesis / digestive tract development / reciprocal meiotic recombination / face morphogenesis / microtubule motor activity / mitotic sister chromatid cohesion / sister chromatid cohesion / negative regulation of interleukin-1 beta production / stem cell population maintenance / mitotic spindle pole / lncRNA binding / dynein complex binding / outflow tract morphogenesis / regulation of DNA replication / mitotic sister chromatid segregation / regulation of embryonic development / positive regulation of interleukin-10 production / somatic stem cell population maintenance / negative regulation of tumor necrosis factor production / developmental growth / mitotic spindle assembly / chromosome, centromeric region / fat cell differentiation / heart morphogenesis / SUMOylation of DNA damage response and repair proteins / beta-tubulin binding / cis-regulatory region sequence-specific DNA binding / protein localization to chromatin / Meiotic synapsis / Resolution of Sister Chromatid Cohesion / condensed nuclear chromosome / meiotic cell cycle / chromosome segregation / promoter-specific chromatin binding / sensory perception of sound / response to radiation / brain development / kinetochore / cognition / nuclear matrix / histone deacetylase binding / spindle pole / Separation of Sister Chromatids / intracellular protein localization / transcription corepressor activity / double-strand break repair / mitotic cell cycle / chromosome / double-stranded DNA binding / midbody / DNA recombination / Estrogen-dependent gene expression / DNA-binding transcription factor binding / negative regulation of neuron apoptotic process / response to hypoxia / chromatin remodeling / intracellular membrane-bounded organelle / protein heterodimerization activity / cell division / DNA repair / apoptotic process / DNA damage response Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
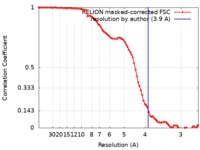
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Shi ZB / Gao H / Bai XC / Yu H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Authors: Zhubing Shi / Haishan Gao / Xiao-Chen Bai / Hongtao Yu /   Abstract: As a ring-shaped adenosine triphosphatase (ATPase) machine, cohesin organizes the eukaryotic genome by extruding DNA loops and mediates sister chromatid cohesion by topologically entrapping DNA. How ...As a ring-shaped adenosine triphosphatase (ATPase) machine, cohesin organizes the eukaryotic genome by extruding DNA loops and mediates sister chromatid cohesion by topologically entrapping DNA. How cohesin executes these fundamental DNA transactions is not understood. Using cryo-electron microscopy (cryo-EM), we determined the structure of human cohesin bound to its loader NIPBL and DNA at medium resolution. Cohesin and NIPBL interact extensively and together form a central tunnel to entrap a 72-base pair DNA. NIPBL and DNA promote the engagement of cohesin's ATPase head domains and ATP binding. The hinge domains of cohesin adopt an "open washer" conformation and dock onto the STAG1 subunit. Our structure explains the synergistic activation of cohesin by NIPBL and DNA and provides insight into DNA entrapment by cohesin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21663.map.gz emd_21663.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21663-v30.xml emd-21663-v30.xml emd-21663.xml emd-21663.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21663_fsc.xml emd_21663_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_21663.png emd_21663.png | 136.4 KB | ||
| Filedesc metadata |  emd-21663.cif.gz emd-21663.cif.gz | 9.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21663 http://ftp.pdbj.org/pub/emdb/structures/EMD-21663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21663 | HTTPS FTP |
-Related structure data
| Related structure data |  6wgeMC  6wg3C  6wg4C  6wg6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21663.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21663.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human Cohesin-NIPBL-DNA Complex without STAG1
| Entire | Name: Human Cohesin-NIPBL-DNA Complex without STAG1 |
|---|---|
| Components |
|
-Supramolecule #1: Human Cohesin-NIPBL-DNA Complex without STAG1
| Supramolecule | Name: Human Cohesin-NIPBL-DNA Complex without STAG1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 820 KDa |
-Macromolecule #1: Structural maintenance of chromosomes protein 1A
| Macromolecule | Name: Structural maintenance of chromosomes protein 1A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 143.484109 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGFLKLIEIE NFKSYKGRQI IGPFQRFTAI IGPNGSGKSN LMDAISFVLG EKTSNLRVKT LRDLIHGAPV GKPAANRAFV SMVYSEEGA EDRTFARVIV GGSSEYKINN KVVQLHEYSE ELEKLGILIK ARNFLVFQGA VESIAMKNPK ERTALFEEIS R SGELAQEY ...String: MGFLKLIEIE NFKSYKGRQI IGPFQRFTAI IGPNGSGKSN LMDAISFVLG EKTSNLRVKT LRDLIHGAPV GKPAANRAFV SMVYSEEGA EDRTFARVIV GGSSEYKINN KVVQLHEYSE ELEKLGILIK ARNFLVFQGA VESIAMKNPK ERTALFEEIS R SGELAQEY DKRKKEMVKA EEDTQFNYHR KKNIAAERKE AKQEKEEADR YQRLKDEVVR AQVQLQLFKL YHNEVEIEKL NK ELASKNK EIEKDKKRMD KVEDELKEKK KELGKMMREQ QQIEKEIKEK DSELNQKRPQ YIKAKENTSH KIKKLEAAKK SLQ NAQKHY KKRKGDMDEL EKEMLSVEKA RQEFEERMEE ESQSQGRDLT LEENQVKKYH RLKEEASKRA ATLAQELEKF NRDQ KADQD RLDLEERKKV ETEAKIKQKL REIEENQKRI EKLEEYITTS KQSLEEQKKL EGELTEEVEM AKRRIDEINK ELNQV MEQL GDARIDRQES SRQQRKAEIM ESIKRLYPGS VYGRLIDLCQ PTQKKYQIAV TKVLGKNMDA IIVDSEKTGR DCIQYI KEQ RGEPETFLPL DYLEVKPTDE KLRELKGAKL VIDVIRYEPP HIKKALQYAC GNALVCDNVE DARRIAFGGH QRHKTVA LD GTLFQKSGVI SGGASDLKAK ARRWDEKAVD KLKEKKERLT EELKEQMKAK RKEAELRQVQ SQAHGLQMRL KYSQSDLE Q TKTRHLALNL QEKSKLESEL ANFGPRINDI KRIIQSRERE MKDLKEKMNQ VEDEVFEEFC REIGVRNIRE FEEEKVKRQ NEIAKKRLEF ENQKTRLGIQ LDFEKNQLKE DQDKVHMWEQ TVKKDENEIE KLKKEEQRHM KIIDETMAQL QDLKNQHLAK KSEVNDKNH EMEEIRKKLG GANKEMTHLQ KEVTAIETKL EQKRSDRHNL LQACKMQDIK LPLSKGTMDD ISQEEGSSQG E DSVSGSQR ISSIYAREAL IEIDYGDLCE DLKDAQAEEE IKQEMNTLQQ KLNEQQSVLQ RIAAPNMKAM EKLESVRDKF QE TSDEFEA ARKRAKKAKQ AFEQIKKERF DRFNACFESV ATNIDEIYKA LSRNSSAQAF LGPENPEEPY LDGINYNCVA PGK RFRPMD NLSGGEKTVA ALALLFAIHS YKPAPFFVLD QIDAALDNTN IGKVANYIKE QSTCNFQAIV ISLKEEFYTK AESL IGVYP EQGDCVISKV LTFDLTKYPD ANPNPNEQ UniProtKB: Structural maintenance of chromosomes protein 1A |
-Macromolecule #2: Structural maintenance of chromosomes protein 3
| Macromolecule | Name: Structural maintenance of chromosomes protein 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 141.770578 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MYIKQVIIQG FRSYRDQTIV DPFSSKHNVI VGRNGSGKSN FFYAIQFVLS DEFSHLRPEQ RLALLHEGTG PRVISAFVEI IFDNSDNRL PIDKEEVSLR RVIGAKKDQY FLDKKMVTKN DVMNLLESAG FSRSNPYYIV KQGKINQMAT APDSQRLKLL R EVAGTRVY ...String: MYIKQVIIQG FRSYRDQTIV DPFSSKHNVI VGRNGSGKSN FFYAIQFVLS DEFSHLRPEQ RLALLHEGTG PRVISAFVEI IFDNSDNRL PIDKEEVSLR RVIGAKKDQY FLDKKMVTKN DVMNLLESAG FSRSNPYYIV KQGKINQMAT APDSQRLKLL R EVAGTRVY DERKEESISL MKETEGKREK INELLKYIEE RLHTLEEEKE ELAQYQKWDK MRRALEYTIY NQELNETRAK LD ELSAKRE TSGEKSRQLR DAQQDARDKM EDIERQVREL KTKISAMKEE KEQLSAERQE QIKQRTKLEL KAKDLQDELA GNS EQRKRL LKERQKLLEK IEEKQKELAE TEPKFNSVKE KEERGIARLA QATQERTDLY AKQGRGSQFT SKEERDKWIK KELK SLDQA INDKKRQIAA IHKDLEDTEA NKEKNLEQYN KLDQDLNEVK ARVEELDRKY YEVKNKKDEL QSERNYLWRE ENAEQ QALA AKREDLEKKQ QLLRAATGKA ILNGIDSINK VLDHFRRKGI NQHVQNGYHG IVMNNFECEP AFYTCVEVTA GNRLFY HIV DSDEVSTKIL MEFNKMNLPG EVTFLPLNKL DVRDTAYPET NDAIPMISKL RYNPRFDKAF KHVFGKTLIC RSMEVST QL ARAFTMDCIT LEGDQVSHRG ALTGGYYDTR KSRLELQKDV RKAEEELGEL EAKLNENLRR NIERINNEID QLMNQMQQ I ETQQRKFKAS RDSILSEMKM LKEKRQQSEK TFMPKQRSLQ SLEASLHAME STRESLKAEL GTDLLSQLSL EDQKRVDAL NDEIRQLQQE NRQLLNERIK LEGIITRVET YLNENLRKRL DQVEQELNEL RETEGGTVLT ATTSELEAIN KRVKDTMARS EDLDNSIDK TEAGIKELQK SMERWKNMEK EHMDAINHDT KELEKMTNRQ GMLLKKKEEC MKKIRELGSL PQEAFEKYQT L SLKQLFRK LEQCNTELKK YSHVNKKALD QFVNFSEQKE KLIKRQEELD RGYKSIMELM NVLELRKYEA IQLTFKQVSK NF SEVFQKL VPGGKATLVM KKGDVEGSQS QDEGEGSGES ERGSGSQSSV PSVDQFTGVG IRVSFTGKQG EMREMQQLSG GQK SLVALA LIFAIQKCDP APFYLFDQID QALDAQHRKA VSDMIMELAV HAQFITTTFR PELLESADKF YGVKFRNKVS HIDV ITAEM AKDFVEDDTT HG UniProtKB: Structural maintenance of chromosomes protein 3 |
-Macromolecule #3: Double-strand-break repair protein rad21 homolog
| Macromolecule | Name: Double-strand-break repair protein rad21 homolog / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.556102 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MFYAHFVLSK RGPLAKIWLA AHWDKKLTKA HVFECNLESS VESIISPKVK MALRTSGHLL LGVVRIYHRK AKYLLADCNE AFIKIKMAF RPGVVDLPEE NREAAYNAIT LPEEFHDFDQ PLPDLDDIDV AQQFSLNQSR VEEITMREEV GNISILQEND F GDFGMDDR ...String: MFYAHFVLSK RGPLAKIWLA AHWDKKLTKA HVFECNLESS VESIISPKVK MALRTSGHLL LGVVRIYHRK AKYLLADCNE AFIKIKMAF RPGVVDLPEE NREAAYNAIT LPEEFHDFDQ PLPDLDDIDV AQQFSLNQSR VEEITMREEV GNISILQEND F GDFGMDDR EIMAEGSAFE DDDMLVSTTT SNLLLESEQS TSNLNEKINH LEYEDQYKDD NFGEGNDGGI LDDKLISNND GG IFDDPPA LSEAGVMLPE QPAHDDMDED DNVSMGGPDS PASVDPVEPM PTMTDQTTLV PNEEEAFALE PIDITVKETK AKR KRKLIV DSVKELDSKT IRAQLSDYSD IVTTLDLAPP TKKLMMWKET GGVEKLFSLP AQPLWNNRLL KLFTRCLTPL VPED LRKRR KGGEADNLDE FLKEFENPEV PREDQQQQHQ QRDVIDEPII EEPSALQESV MEASRTNIDE SAMPPPPPQG VKRKA GQID PEPVMPPQQV EQMEIPPVEL PPEEPPNICQ LIPELELLPE KEKEKEKEKE DDEEEEDEDA SGGDQDQEER RWNKRT QQM LHGLQRALAK TGAESISLLE LCRNTNRKQA AAKFYSFLVL KKQQAIELTQ EEPYSDIIAT PGPRFHII UniProtKB: Double-strand-break repair protein rad21 homolog |
-Macromolecule #4: Nipped-B-like protein
| Macromolecule | Name: Nipped-B-like protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 186.77825 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: PSLSEVARKM KKKEKQKKRK AYEPKLTPEE MMDSSTFKRF TASIENILDN LEDMDFTAFG DDDEIPQELL LGKHQLNELG SESAKIKAM GIMDKLSTDK TVKVLNILEK NIQDGSKLST LLNHNNDTEE EERLWRDLIM ERVTKSADAC LTTINIMTSP N MPKAVYIE ...String: PSLSEVARKM KKKEKQKKRK AYEPKLTPEE MMDSSTFKRF TASIENILDN LEDMDFTAFG DDDEIPQELL LGKHQLNELG SESAKIKAM GIMDKLSTDK TVKVLNILEK NIQDGSKLST LLNHNNDTEE EERLWRDLIM ERVTKSADAC LTTINIMTSP N MPKAVYIE DVIERVIQYT KFHLQNTLYP QYDPVYRLDP HGGGLLSSKA KRAKCSTHKQ RVIVMLYNKV CDIVSSLSEL LE IQLLTDT TILQVSSMGI TPFFVENVSE LQLCAIKLVT AVFSRYEKHR QLILEEIFTS LARLPTSKRS LRNFRLNSSD MDG EPMYIQ MVTALVLQLI QCVVHLPSSE KDSNAEEDSN KKIDQDVVIT NSYETAMRTA QNFLSIFLKK CGSKQGEEDY RPLF ENFVQ DLLSTVNKPE WPAAELLLSL LGRLLVHQFS NKSTEMALRV ASLDYLGTVA ARLRKDAVTS KMDQGSIERI LKQVS GGED EIQQLQKALL DYLDENTETD PSLVFSRKFY IAQWFRDTTL ETEKAMKSQK DEESSEGTHH AKEIETTGQI MHRAEN RKK FLRSIIKTTP SQFSTLKMNS DTVDYDDACL IVRYLASMRP FAQSFDIYLT QILRVLGENA IAVRTKAMKC LSEVVAV DP SILARLDMQR GVHGRLMDNS TSVREAAVEL LGRFVLCRPQ LAEQYYDMLI ERILDTGISV RKRVIKILRD ICIEQPTF P KITEMCVKMI RRVNDEEGIK KLVNETFQKL WFTPTPHNDK EAMTRKILNI TDVVAACRDT GYDWFEQLLQ NLLKSEEDS SYKPVKKACT QLVDNLVEHI LKYEESLADS DNKGVNSGRL VACITTLFLF SKIRPQLMVK HAMTMQPYLT TKCSTQNDFM VICNVAKIL ELVVPLMEHP SETFLATIEE DLMKLIIKYG MTVVQHCVSC LGAVVNKVTQ NFKFVWACFN RYYGAISKLK S QHQEDPNN TSLLTNKPAL LRSLFTVGAL CRHFDFDLED FKGNSKVNIK DKVLELLMYF TKHSDEEVQT KAIIGLGFAF IQ HPSLMFE QEVKNLYNNI LSDKNSSVNL KIQVLKNLQT YLQEEDTRMQ QADRDWKKVA KQEDLKEMGD VSSGMSSSIM QLY LKQVLE AFFHTQSSVR HFALNVIALT LNQGLIHPVQ CVPYLIAMGT DPEPAMRNKA DQQLVEIDKK YAGFIHMKAV AGMK MSYQV QQAINTCLKD PVRGFRQDES SSALCSHLYS MIRGNRQHRR AFLISLLNLF DDTAKTDVTM LLYIADNLAC FPYQT QEEP LFIMHHIDIT LSVSGSNLLQ SFKESMVKDK RKERKSSPSK ENESSDSEEE VSRPRKSRKR VDSDSDSDSE DDINSV MKC LPENSAPLIE FANVSQGILL LLMLKQHLKN LCGFSDSKIQ KYSPSESAKV YDKAINRKTG VHFHPKQTLD FLRSDMA NS KITEEVKRSI VKQYLDFKLL MEHLDPDEEE EEGEVSASTN ARNKAITSLL GGGSPKNNTA AETEDDESDG EDRGGGTS G SLRRSKRNSD STELAAQMNE SVDVMDVIAI CCPKYKDRPQ IARVVQKTSS GFSVQWMAGS YSGSWTEAKR RDGRKLVPW VDTIKESDII YKKIALTSAN KLTNKVVQTL RSLYAAKDGT SS UniProtKB: Nipped-B-like protein |
-Macromolecule #5: DNA (43-MER)
| Macromolecule | Name: DNA (43-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.422931 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA) (DA)(DA)(DA) |
-Macromolecule #6: DNA (43-MER)
| Macromolecule | Name: DNA (43-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.035333 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT) (DT)(DT)(DT) |
-Macromolecule #7: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 7 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 5796 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6wge: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)