[English] 日本語
 Yorodumi
Yorodumi- EMDB-21601: Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21601 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | |||||||||||||||||||||
 Map data Map data | Sharpened map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Glycosyltransferase / membrane protein / nanodisc / acyl carrier protein | |||||||||||||||||||||
| Function / homology | Alpha-(1->3)-arabinofuranosyltransferase / Alpha-(1->3)-arabinofuranosyltransferase, N-terminal GT-C domain / Coagulation factors 5/8 type C domain (FA58C) profile. / Coagulation factor 5/8 C-terminal domain / Galactose-binding-like domain superfamily / transferase activity / membrane / DUF3367 domain-containing protein Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Mycobacteroides abscessus (bacteria) Mycobacteroides abscessus (bacteria) | |||||||||||||||||||||
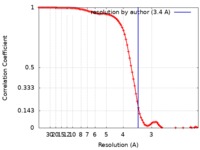
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||||||||
 Authors Authors | Tan YZ / Zhang L | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria. Authors: Yong Zi Tan / Lei Zhang / José Rodrigues / Ruixiang Blake Zheng / Sabrina I Giacometti / Ana L Rosário / Brian Kloss / Venkata P Dandey / Hui Wei / Richard Brunton / Ashleigh M Raczkowski ...Authors: Yong Zi Tan / Lei Zhang / José Rodrigues / Ruixiang Blake Zheng / Sabrina I Giacometti / Ana L Rosário / Brian Kloss / Venkata P Dandey / Hui Wei / Richard Brunton / Ashleigh M Raczkowski / Diogo Athayde / Maria João Catalão / Madalena Pimentel / Oliver B Clarke / Todd L Lowary / Margarida Archer / Michael Niederweis / Clinton S Potter / Bridget Carragher / Filippo Mancia /     Abstract: Mycobacterium tuberculosis causes tuberculosis, a disease that kills over 1 million people each year. Its cell envelope is a common antibiotic target and has a unique structure due, in part, to two ...Mycobacterium tuberculosis causes tuberculosis, a disease that kills over 1 million people each year. Its cell envelope is a common antibiotic target and has a unique structure due, in part, to two lipidated polysaccharides-arabinogalactan and lipoarabinomannan. Arabinofuranosyltransferase D (AftD) is an essential enzyme involved in assembling these glycolipids. We present the 2.9-Å resolution structure of M. abscessus AftD, determined by single-particle cryo-electron microscopy. AftD has a conserved GT-C glycosyltransferase fold and three carbohydrate-binding modules. Glycan array analysis shows that AftD binds complex arabinose glycans. Additionally, AftD is non-covalently complexed with an acyl carrier protein (ACP). 3.4- and 3.5-Å structures of a mutant with impaired ACP binding reveal a conformational change, suggesting that ACP may regulate AftD function. Mutagenesis experiments using a conditional knockout constructed in M. smegmatis confirm the essentiality of the putative active site and the ACP binding for AftD function. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21601.map.gz emd_21601.map.gz | 55.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21601-v30.xml emd-21601-v30.xml emd-21601.xml emd-21601.xml | 33.4 KB 33.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21601_fsc.xml emd_21601_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_21601.jpg emd_21601.jpg emd_21601.png emd_21601.png | 457.5 KB 137 KB | ||
| Masks |  emd_21601_msk_1.map emd_21601_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21601.cif.gz emd-21601.cif.gz | 7.8 KB | ||
| Others |  emd_21601_additional_1.map.gz emd_21601_additional_1.map.gz emd_21601_additional_2.map.gz emd_21601_additional_2.map.gz emd_21601_additional_3.map.gz emd_21601_additional_3.map.gz emd_21601_additional_4.map.gz emd_21601_additional_4.map.gz emd_21601_additional_5.map.gz emd_21601_additional_5.map.gz emd_21601_half_map_1.map.gz emd_21601_half_map_1.map.gz emd_21601_half_map_2.map.gz emd_21601_half_map_2.map.gz | 16.5 MB 2.6 MB 8.5 MB 7.3 MB 108.5 KB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21601 http://ftp.pdbj.org/pub/emdb/structures/EMD-21601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21601 | HTTPS FTP |
-Related structure data
| Related structure data |  6wbyMC  6w98C  6wbxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10391 (Title: Single-Particle Cryo-EM of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S EMPIAR-10391 (Title: Single-Particle Cryo-EM of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389SData size: 2.3 TB Data #1: Unaligned and compressed multi-frame movies [micrographs - multiframe] Data #2: Aligned and dose-weighted micrographs [micrographs - single frame] Data #3: Final Particle Stacks with Refined Euler Angles and Shifts (Overall and Focused Refined on CBM3) [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21601.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21601.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0605 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
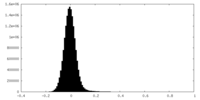
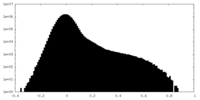
-Mask #1
| File |  emd_21601_msk_1.map emd_21601_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
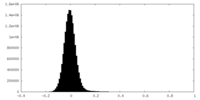
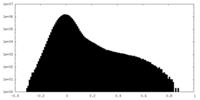
-Additional map: Raw map
| File | emd_21601_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local resolution map
| File | emd_21601_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3DFSC
| File | emd_21601_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFSC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3DFSC - Thresholded
| File | emd_21601_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFSC - Thresholded | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3DFSC - Thresholded, Binarized
| File | emd_21601_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFSC - Thresholded, Binarized | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_21601_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_21601_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mutant R1389S Class 2 Mycobacterial Arabinofuranosyltransferase A...
| Entire | Name: Mutant R1389S Class 2 Mycobacterial Arabinofuranosyltransferase AftD Complexed with Acyl Carrier Protein |
|---|---|
| Components |
|
-Supramolecule #1: Mutant R1389S Class 2 Mycobacterial Arabinofuranosyltransferase A...
| Supramolecule | Name: Mutant R1389S Class 2 Mycobacterial Arabinofuranosyltransferase AftD Complexed with Acyl Carrier Protein type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Mycobacteroides abscessus (bacteria) Mycobacteroides abscessus (bacteria) |
| Molecular weight | Theoretical: 5 MDa |
-Macromolecule #1: DUF3367 domain-containing protein
| Macromolecule | Name: DUF3367 domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacteroides abscessus (bacteria) Mycobacteroides abscessus (bacteria) |
| Molecular weight | Theoretical: 152.818641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDDDDKH HHHHHHHHHE NLYFQSYVMT YRLDSSALSR RWLAVAAAVS LLLTFSQSPG QISPDTKLDL AINPLRFAAR ALNLWSSDL PFGQAQNQAY GYLFPHGAFF SLGHLLGVPA WVTQRLWWAL LIVAGFWGLI RVAEALGIGT RGSRIIAAVA F ALSPRVLT ...String: MDYKDDDDKH HHHHHHHHHE NLYFQSYVMT YRLDSSALSR RWLAVAAAVS LLLTFSQSPG QISPDTKLDL AINPLRFAAR ALNLWSSDL PFGQAQNQAY GYLFPHGAFF SLGHLLGVPA WVTQRLWWAL LIVAGFWGLI RVAEALGIGT RGSRIIAAVA F ALSPRVLT TLGAISSETL PMMLAPWVLL PLILTFQGRM SPRRAAALSA VAVALMGAVN AVATALACGV AVIWWLAHRP NR TWWRFTA WWIPCLALAS TWWIVALLIF GKISPKFLDF IESSGVTTQW TSLTEVLRGT DSWTPFVAPT ATAGSSLVTQ SAM VIATTM LAAAGMAGLA MRGMPARGRL VAVLLIGLVL LTAGYTGALG SPIAQQIQFF LDDGGTPLRN VHKLEPLIRL PLIL GLAHA LSRIPLPASV PVRQWLSALA RPERNRAVAF AIVLLVALAA STSLAWTGRL VPRGGFDAIP GYWNDTAHWL ADHDT GGRA LVVPGAPFAI QTWGLTRDEP LQALGQTPWG VRDSIPLTPP ETIRAIDSVQ QLFAAGRPSD GLADTLREQG ISYLVV RND LDPDTSRSAR PILVHHTIEG SPGLTKVAQF GDPVGAGAVE GFVADSDLRP QYPAVEIYAV GANDHDGEPY FTDIDTM PR VAGGPEALLR LNERRRQLNE PPLGPSLLAT DAAQAGLRPG PAVVTDTPLA RETDYGRVDD HSSAIRAPGD KRRTFNRV P DYPATGVPLV NGSWTGGTIT ASSSASDSTA LPNVAPGTST AAAIDRDNAT SWVSSSLEAA LGQWIRIDLD RPITNAILT VTPSATALGA QVRRLEVETD NGTTSVRFDE PGQPLNIALR PGETTWVKVT ATGTDDGTSG VQFGVTELSL TQYDAAGFAH TVDLRHSAT VPPPPAGDNP LGWDLGSPLQ GRSGCAPSPQ RLRCAATLSL APEEPGTFIR TLTVPQPVSL TPRLWVRARP G PQLRDLIQ QPGTTVATGD SDVIDPQGSS YAATDGDPGT VWTAPQDSVQ RLHLPSLVIK LPKPTAIGAI RLRPSRTEVP AH PKQVAIN LGDGPQLRSI DPKADVTELA LHPSITDTIT VTVTDWTDII DRTALGFDQL KPPGIAEVIA LDADHRPIAP ADN AANSKR KITIGCNRGP ILALAGRFVP MSITATVREL LDGTVIQATP CDTSPIATGA GIQDVTVNPS QQFIVDGVQL TAAA TEPAS ATMTVAPKGA WGPDRREVTA EPSAHERVLA VPESINPGWA ARDAQGHLLT PVRVNGWQQG WVLPAGDGGK ITLTF GLNT WYRAGLFGGL ALLPILACLA LLPARGRTTL PPVAPWCAGP AAGVAVLAAL TAISGISGMA VGLAALAFKV WTRWPL RAV TAAGVYLAGG SLLLAGAALS RHPWRSVGGY TGHSWWIQLL ALISVASVAL AAVSLPSRRC WKRRSASREG DSTSA UniProtKB: DUF3367 domain-containing protein |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solution was filtered and degassed. | |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 298 K / Instrument: LEICA EM GP | |||||||||
| Details | Protein was incorporated into lipid nanodiscs. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-90 / Number grids imaged: 1 / Number real images: 4886 / Average exposure time: 13.05 sec. / Average electron dose: 96.78 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-6wby: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)