[English] 日本語
 Yorodumi
Yorodumi- EMDB-21397: Porcine epidemic diarrhea virus (PEDV) spike protein, N-terminal ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21397 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Porcine epidemic diarrhea virus (PEDV) spike protein, N-terminal domain deletion, expressed in Sf9 cells and negatively stained | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus | |||||||||
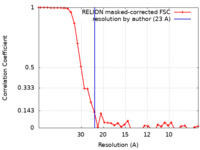
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Kirchdoerfer RN / Martini O / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Structure and immune recognition of the porcine epidemic diarrhea virus spike protein. Authors: Robert N Kirchdoerfer / Mahesh Bhandari / Olnita Martini / Leigh M Sewall / Sandhya Bangaru / Kyoung-Jin Yoon / Andrew B Ward /  Abstract: Porcine epidemic diarrhea virus (PEDV) is an alphacoronavirus responsible for significant morbidity and mortality in pigs. A key determinant of viral tropism and entry, the PEDV spike protein is a ...Porcine epidemic diarrhea virus (PEDV) is an alphacoronavirus responsible for significant morbidity and mortality in pigs. A key determinant of viral tropism and entry, the PEDV spike protein is a key target for the host antibody response and a good candidate for a protein-based vaccine immunogen. We used electron microscopy to evaluate the PEDV spike structure, as well as pig polyclonal antibody responses to viral infection. The structure of the PEDV spike reveals a configuration similar to that of HuCoV-NL63. Several PEDV protein-protein interfaces are mediated by non-protein components, including a glycan at Asn264 and two bound palmitoleic acid molecules. The polyclonal antibody response to PEDV infection shows a dominance of epitopes in the S1 region. This structural and immune characterization provides insights into coronavirus spike stability determinants and explores the immune landscape of viral spike proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
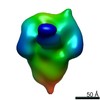
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21397.map.gz emd_21397.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21397-v30.xml emd-21397-v30.xml emd-21397.xml emd-21397.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21397_fsc.xml emd_21397_fsc.xml | 3.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_21397.png emd_21397.png | 59.6 KB | ||
| Masks |  emd_21397_msk_1.map emd_21397_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Others |  emd_21397_half_map_1.map.gz emd_21397_half_map_1.map.gz emd_21397_half_map_2.map.gz emd_21397_half_map_2.map.gz | 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21397 http://ftp.pdbj.org/pub/emdb/structures/EMD-21397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21397 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21397 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21397.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21397.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
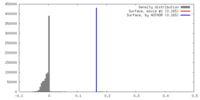
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21397_msk_1.map emd_21397_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
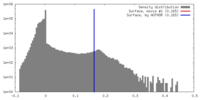
| Density Histograms |
-Half map: #1
| File | emd_21397_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
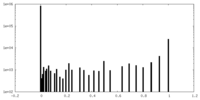
| Density Histograms |
-Half map: #2
| File | emd_21397_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
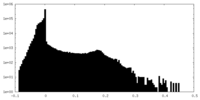
| Density Histograms |
- Sample components
Sample components
-Entire : Porcine epidemic diarrhea virus spike protein with a deletion of ...
| Entire | Name: Porcine epidemic diarrhea virus spike protein with a deletion of the N-terminal domain 0 |
|---|---|
| Components |
|
-Supramolecule #1: Porcine epidemic diarrhea virus spike protein with a deletion of ...
| Supramolecule | Name: Porcine epidemic diarrhea virus spike protein with a deletion of the N-terminal domain 0 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinantly expressed in Sf9 cells using baculovirus |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus / Strain: 13-019349 Porcine epidemic diarrhea virus / Strain: 13-019349 |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 600 KDa |
-Macromolecule #1: Porcine epidemic diarrhea virus (PEDV) spike protein
| Macromolecule | Name: Porcine epidemic diarrhea virus (PEDV) spike protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Recombinant expression | Organism:  |
| Sequence | String: LPQDVTRCSA NTNFRTANCI GYAANVFATE PNGHIPEGFS FN NWFLLSN DSTLVHGKVV SNQPLLVNCL LAIPKIYGLG QFFSFNQTID GVCNGAAVQR APE ALRFNI NDISVILAEG SIVLHTALGT NFSFVCSNSS NPHLATFAIP LGATQVPYYC FLKV DTYNS ...String: LPQDVTRCSA NTNFRTANCI GYAANVFATE PNGHIPEGFS FN NWFLLSN DSTLVHGKVV SNQPLLVNCL LAIPKIYGLG QFFSFNQTID GVCNGAAVQR APE ALRFNI NDISVILAEG SIVLHTALGT NFSFVCSNSS NPHLATFAIP LGATQVPYYC FLKV DTYNS TVYKFLAVLP PTVREIVITK YGDVYVNGFG YLHLGLLDAV TINFTGHGTD DDVSG FWTI ASTNFVDALI EVQGTAIQRI LYCDDPVSQL KCSQVAFDLD DGFYTISSRN LLSHEQ PIS FVTLPSFNDH SFVNITVSAS FGGHSGANLI ASDTTINGFS SFCVDTRQFT ISLFYNV TN SYGYVSKSQD SNCPFTLQSV NDYLSFSKFC VSTSLLASAC TIDLFGYPEF GSGVKFTS L YFQFTKGELI TGTPKPFEGV TDVSFMTLDV CTKYTIYGFK GEGIITLTNS SFLAGVYYT SDSGQLLAFK NVTSGAVYSV TPCSFSEQAA YVDDDIVGVI SSLSSSTFNS TRELPGFFYH SNDGSNCTE PVLVYSNIGV CKSGSIGYVP SQSGQVKIAP TVTGNISIPT NFSMSIRTEY L QLYNTPVS VDCATYVCNG NSRCKQLLTQ YTAACKTIES ALQLSARLES VEVNSMLTIS DE ALQLATI SSFNGDGYNF TNVLGVSVYD PASGRVVQKR SFIEDLLFNK VVTNGLGTVD EDY KRCSNG RSVADLVCAQ YYSGVMVLPG VVDAEKLHMY SASLIGGMVL GGFTSAAALP FSYA VQARL NYLALQTDVL QRNQQLLAES FNSAIGNITS AFESVKEAIS QTSKGLNTVA HALTK VQEV VNSQGAALTQ LTVQLQHNFQ AISSSIDDIY SRLDILSADA QVDRLITGRL SALNAF VAQ TLTKYTEVQA SRKLAQQKVN ECVKSQSQRY GFCGGDGEHI FSLVQAAPQG LLFLHTV LV PSDFVDVIAI AGLCVNDEIA LTLREPGLVL FTHELQNHTA TEYFVSSRRM FEPRKPTV S DFVQIESCVV TYVNLTRDQL PDVIPDYIDV NKTLDEILAS LPNRTGPSLP LDVFNATYL NLTGEIADLE QRSESLRNTT EELQSLIYNI NNTLVDLEWL NRVETGSGYI PEAPRDGQAY VRKDGEWVL LSTFLENLYF QGGHHHHHHA WSHPQFEK |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||
| Staining | Type: NEGATIVE / Material: Uranyl formate | ||||||
| Grid | Support film - Material: CARBON / Support film - topology: CONTINUOUS / Details: unspecified |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-64 / Number grids imaged: 1 / Average exposure time: 0.6 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)