[English] 日本語
 Yorodumi
Yorodumi- EMDB-21391: Cryo-EM structure of porcine epidemic diarrhea virus (PEDV) spike... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21391 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of porcine epidemic diarrhea virus (PEDV) spike protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycoprotein / surface / receptor-binding / membrane fusion / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Kirchdoerfer RN / Ward AB | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Structure and immune recognition of the porcine epidemic diarrhea virus spike protein. Authors: Robert N Kirchdoerfer / Mahesh Bhandari / Olnita Martini / Leigh M Sewall / Sandhya Bangaru / Kyoung-Jin Yoon / Andrew B Ward /  Abstract: Porcine epidemic diarrhea virus (PEDV) is an alphacoronavirus responsible for significant morbidity and mortality in pigs. A key determinant of viral tropism and entry, the PEDV spike protein is a ...Porcine epidemic diarrhea virus (PEDV) is an alphacoronavirus responsible for significant morbidity and mortality in pigs. A key determinant of viral tropism and entry, the PEDV spike protein is a key target for the host antibody response and a good candidate for a protein-based vaccine immunogen. We used electron microscopy to evaluate the PEDV spike structure, as well as pig polyclonal antibody responses to viral infection. The structure of the PEDV spike reveals a configuration similar to that of HuCoV-NL63. Several PEDV protein-protein interfaces are mediated by non-protein components, including a glycan at Asn264 and two bound palmitoleic acid molecules. The polyclonal antibody response to PEDV infection shows a dominance of epitopes in the S1 region. This structural and immune characterization provides insights into coronavirus spike stability determinants and explores the immune landscape of viral spike proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21391.map.gz emd_21391.map.gz | 78.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21391-v30.xml emd-21391-v30.xml emd-21391.xml emd-21391.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21391_fsc.xml emd_21391_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_21391.png emd_21391.png | 182.9 KB | ||
| Masks |  emd_21391_msk_1.map emd_21391_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21391.cif.gz emd-21391.cif.gz | 7.2 KB | ||
| Others |  emd_21391_half_map_1.map.gz emd_21391_half_map_1.map.gz emd_21391_half_map_2.map.gz emd_21391_half_map_2.map.gz | 65.4 MB 65.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21391 http://ftp.pdbj.org/pub/emdb/structures/EMD-21391 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21391 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21391 | HTTPS FTP |
-Related structure data
| Related structure data |  6vv5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21391.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21391.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21391_msk_1.map emd_21391_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: First half map
| File | emd_21391_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Second half map
| File | emd_21391_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Porcine epidemic diarrhea virus spike protein
| Entire | Name: Porcine epidemic diarrhea virus spike protein |
|---|---|
| Components |
|
-Supramolecule #1: Porcine epidemic diarrhea virus spike protein
| Supramolecule | Name: Porcine epidemic diarrhea virus spike protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Recombinantly expressed in insect cells using a baculovirus vector. |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus / Strain: 13-019349 Porcine epidemic diarrhea virus / Strain: 13-019349 |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Porcine epidemic diarrhea virus Porcine epidemic diarrhea virus |
| Molecular weight | Theoretical: 148.463656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LPQDVTRCSA NTNFRRFFSK FNVQAPAVVV LGGYLPIGEN QGVNSTWYCA GQHPTASGVH GIFVSHIRGG HGFEIGISQE PFDPSGYQL YLHKATNGNT NATARLRICQ FPSIKTLGPT ANNDVTTGRN CLFNKAIPAH MSEHSVVGIT WDNDRVTVFS D KIYYFYFK ...String: LPQDVTRCSA NTNFRRFFSK FNVQAPAVVV LGGYLPIGEN QGVNSTWYCA GQHPTASGVH GIFVSHIRGG HGFEIGISQE PFDPSGYQL YLHKATNGNT NATARLRICQ FPSIKTLGPT ANNDVTTGRN CLFNKAIPAH MSEHSVVGIT WDNDRVTVFS D KIYYFYFK NDWSRVATKC YNSGGCAMQY VYEPTYYMLN VTSAGEDGIS YQPCTANCIG YAANVFATEP NGHIPEGFSF NN WFLLSND STLVHGKVVS NQPLLVNCLL AIPKIYGLGQ FFSFNQTIDG VCNGAAVQRA PEALRFNIND ISVILAEGSI VLH TALGTN FSFVCSNSSN PHLATFAIPL GATQVPYYCF LKVDTYNSTV YKFLAVLPPT VREIVITKYG DVYVNGFGYL HLGL LDAVT INFTGHGTDD DVSGFWTIAS TNFVDALIEV QGTAIQRILY CDDPVSQLKC SQVAFDLDDG FYTISSRNLL SHEQP ISFV TLPSFNDHSF VNITVSASFG GHSGANLIAS DTTINGFSSF CVDTRQFTIS LFYNVTNSYG YVSKSQDSNC PFTLQS VND YLSFSKFCVS TSLLASACTI DLFGYPEFGS GVKFTSLYFQ FTKGELITGT PKPFEGVTDV SFMTLDVCTK YTIYGFK GE GIITLTNSSF LAGVYYTSDS GQLLAFKNVT SGAVYSVTPC SFSEQAAYVD DDIVGVISSL SSSTFNSTRE LPGFFYHS N DGSNCTEPVL VYSNIGVCKS GSIGYVPSQS GQVKIAPTVT GNISIPTNFS MSIRTEYLQL YNTPVSVDCA TYVCNGNSR CKQLLTQYTA ACKTIESALQ LSARLESVEV NSMLTISDEA LQLATISSFN GDGYNFTNVL GVSVYDPASG RVVQKRSFIE DLLFNKVVT NGLGTVDEDY KRCSNGRSVA DLVCAQYYSG VMVLPGVVDA EKLHMYSASL IGGMVLGGFT SAAALPFSYA V QARLNYLA LQTDVLQRNQ QLLAESFNSA IGNITSAFES VKEAISQTSK GLNTVAHALT KVQEVVNSQG AALTQLTVQL QH NFQAISS SIDDIYSRLD ILSADAQVDR LITGRLSALN AFVAQTLTKY TEVQASRKLA QQKVNECVKS QSQRYGFCGG DGE HIFSLV QAAPQGLLFL HTVLVPSDFV DVIAIAGLCV NDEIALTLRE PGLVLFTHEL QNHTATEYFV SSRRMFEPRK PTVS DFVQI ESCVVTYVNL TRDQLPDVIP DYIDVNKTLD EILASLPNRT GPSLPLDVFN ATYLNLTGEI ADLEQRSESL RNTTE ELQS LIYNINNTLV DLEWLNRVET GSGYIPEAPR DGQAYVRKDG EWVLLSTFLE NLYFQGGHHH HHHAWSHPQF EK UniProtKB: Spike glycoprotein |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #6: PALMITOLEIC ACID
| Macromolecule | Name: PALMITOLEIC ACID / type: ligand / ID: 6 / Number of copies: 6 / Formula: PAM |
|---|---|
| Molecular weight | Theoretical: 254.408 Da |
| Chemical component information |  ChemComp-PAM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | ||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Temperature | Max: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-64 / Number grids imaged: 1 / Average exposure time: 12.8 sec. / Average electron dose: 54.9 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












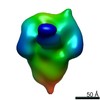











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)