[English] 日本語
 Yorodumi
Yorodumi- EMDB-21243: Structure of a D2 dopamine receptor-G-protein complex in a lipid ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21243 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane | |||||||||
 Map data Map data | Catecholamine receptor structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Dopamine / Dopamine receptor / GPCR / G protein / Parkinson's disease / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of dopamine receptor signaling pathway / positive regulation of dopamine uptake involved in synaptic transmission / negative regulation of dephosphorylation / positive regulation of glial cell-derived neurotrophic factor production / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / nervous system process involved in regulation of systemic arterial blood pressure / response to histamine / negative regulation of circadian sleep/wake cycle, sleep / regulation of synapse structural plasticity ...negative regulation of dopamine receptor signaling pathway / positive regulation of dopamine uptake involved in synaptic transmission / negative regulation of dephosphorylation / positive regulation of glial cell-derived neurotrophic factor production / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / nervous system process involved in regulation of systemic arterial blood pressure / response to histamine / negative regulation of circadian sleep/wake cycle, sleep / regulation of synapse structural plasticity / regulation of locomotion involved in locomotory behavior / neuron-neuron synaptic transmission / negative regulation of dopamine secretion / Extra-nuclear estrogen signaling / adenohypophysis development / negative regulation of cellular response to hypoxia / Adenylate cyclase inhibitory pathway / hyaloid vascular plexus regression / adenylate cyclase-inhibiting dopamine receptor signaling pathway / cerebral cortex GABAergic interneuron migration / response to inactivity / orbitofrontal cortex development / regulation of potassium ion transport / Dopamine receptors / negative regulation of neuron migration / dopamine binding / branching morphogenesis of a nerve / regulation of dopamine uptake involved in synaptic transmission / phospholipase C-activating dopamine receptor signaling pathway / positive regulation of growth hormone secretion / peristalsis / heterotrimeric G-protein binding / drinking behavior / G protein-coupled receptor complex / grooming behavior / behavioral response to ethanol / positive regulation of renal sodium excretion / auditory behavior / striatum development / positive regulation of G protein-coupled receptor signaling pathway / dopaminergic synapse / positive regulation of multicellular organism growth / G protein-coupled receptor internalization / negative regulation of synaptic transmission / non-motile cilium / GTPase activating protein binding / heterocyclic compound binding / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / response to iron ion / negative regulation of synaptic transmission, glutamatergic / positive regulation of urine volume / adult walking behavior / G alpha (i) signalling events / arachidonate secretion / ciliary membrane / negative regulation of cytosolic calcium ion concentration / positive regulation of neuroblast proliferation / response to morphine / regulation of synaptic transmission, GABAergic / neurotransmitter receptor localization to postsynaptic specialization membrane / temperature homeostasis / positive regulation of cytokinesis / pigmentation / dopamine metabolic process / dopamine uptake involved in synaptic transmission / regulation of dopamine secretion / positive regulation of receptor internalization / cellular response to ethanol / response to light stimulus / lateral plasma membrane / associative learning / neuroblast proliferation / G-protein alpha-subunit binding / negative regulation of protein secretion / endocytic vesicle / prepulse inhibition / potassium channel regulator activity / long-term memory / sperm flagellum / response to axon injury / viral release from host cell by cytolysis / postsynaptic modulation of chemical synaptic transmission / negative regulation of blood pressure / regulation of sodium ion transport / behavioral response to cocaine / positive regulation of protein localization to cell cortex / cellular response to retinoic acid / T cell migration / synapse assembly / D2 dopamine receptor binding / peptidoglycan catabolic process / response to prostaglandin E / release of sequestered calcium ion into cytosol / ionotropic glutamate receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / presynaptic modulation of chemical synaptic transmission / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
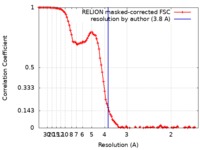
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Yin J / Chen KM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane. Authors: Jie Yin / Kuang-Yui M Chen / Mary J Clark / Mahdi Hijazi / Punita Kumari / Xiao-Chen Bai / Roger K Sunahara / Patrick Barth / Daniel M Rosenbaum /   Abstract: The D2 dopamine receptor (DRD2) is a therapeutic target for Parkinson's disease and antipsychotic drugs. DRD2 is activated by the endogenous neurotransmitter dopamine and synthetic agonist drugs such ...The D2 dopamine receptor (DRD2) is a therapeutic target for Parkinson's disease and antipsychotic drugs. DRD2 is activated by the endogenous neurotransmitter dopamine and synthetic agonist drugs such as bromocriptine, leading to stimulation of G and inhibition of adenylyl cyclase. Here we used cryo-electron microscopy to elucidate the structure of an agonist-bound activated DRD2-G complex reconstituted into a phospholipid membrane. The extracellular ligand-binding site of DRD2 is remodelled in response to agonist binding, with conformational changes in extracellular loop 2, transmembrane domain 5 (TM5), TM6 and TM7, propagating to opening of the intracellular G-binding site. The DRD2-G structure represents, to our knowledge, the first experimental model of a G-protein-coupled receptor-G-protein complex embedded in a phospholipid bilayer, which serves as a benchmark to validate the interactions seen in previous detergent-bound structures. The structure also reveals interactions that are unique to the membrane-embedded complex, including helix 8 burial in the inner leaflet, ordered lysine and arginine side chains in the membrane interfacial regions, and lipid anchoring of the G protein in the membrane. Our model of the activated DRD2 will help to inform the design of subtype-selective DRD2 ligands for multiple human central nervous system disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21243.map.gz emd_21243.map.gz | 78.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21243-v30.xml emd-21243-v30.xml emd-21243.xml emd-21243.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21243_fsc.xml emd_21243_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_21243.png emd_21243.png | 18.8 KB | ||
| Filedesc metadata |  emd-21243.cif.gz emd-21243.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21243 http://ftp.pdbj.org/pub/emdb/structures/EMD-21243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21243 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21243 | HTTPS FTP |
-Related structure data
| Related structure data |  6vmsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21243.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21243.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Catecholamine receptor structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : D2 dopamine receptor-G protein complex
| Entire | Name: D2 dopamine receptor-G protein complex |
|---|---|
| Components |
|
-Supramolecule #1: D2 dopamine receptor-G protein complex
| Supramolecule | Name: D2 dopamine receptor-G protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.382047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDAA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDAA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.41693 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL ...String: MSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKLI IWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC RFLDDNQIVT S SGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD INAICFFPNG NA FATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAGHDNRVSC LGV TDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.137474 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH GGASNNTASI AQARKLVEQL KMEANIDRIK VSKAAADLMA YCEAHAKEDP LLTPVPASEN PFREKKFFCA IL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.784896 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAHHHHHH HH |
-Macromolecule #5: Endolysin,D(2) dopamine receptor,D(2) dopamine receptor
| Macromolecule | Name: Endolysin,D(2) dopamine receptor,D(2) dopamine receptor type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: lysozyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 51.384059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDANI FEMLRIDEGL RLKIYKNTEG YYTIGIGHLL TKSPSLNAAK SELDKAIGRN TNGVITKDEA EKLFNQDVDA AVRGILRNA KLKPVYDSLD AVRRAALINM VFQMGETGVA GFTNSLRMLQ QKRWDEAAVN LAKSRWYNQT PNRAKRVITT F RTGTWDAY ...String: DYKDDDDANI FEMLRIDEGL RLKIYKNTEG YYTIGIGHLL TKSPSLNAAK SELDKAIGRN TNGVITKDEA EKLFNQDVDA AVRGILRNA KLKPVYDSLD AVRRAALINM VFQMGETGVA GFTNSLRMLQ QKRWDEAAVN LAKSRWYNQT PNRAKRVITT F RTGTWDAY AADRPHYNYY ATLLTLLIAV IVFGNVLVCM AVSREKALQT TTNYLIVSLA VADLLVATLV MPWVVYLEVV GE WKFSRIH CDIFVTLDVM MCTASILNLC AISIDRYTAV AMPMLYNTRY SSKRRVTVMI SIVWVLSFTI SCPLLFGLNN ADQ NECIIA NPAFVVYSSI VSFYVPFIVI LLVYIKIYIV LRRRRKLVNT NRKLSQQKEK KATQLLAIYL GLFIICWLPF FITH ILNIH CDCNIPPVLY SAFTWLGYVN SAINPIIYTT FNIEFRKAFL KILHC UniProtKB: Endolysin, D(2) dopamine receptor, D(2) dopamine receptor |
-Macromolecule #6: bromoergocryptine
| Macromolecule | Name: bromoergocryptine / type: ligand / ID: 6 / Number of copies: 1 / Formula: 08Y |
|---|---|
| Molecular weight | Theoretical: 654.594 Da |
| Chemical component information | 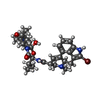 ChemComp-08Y: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)