+Search query
-Structure paper
| Title | Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane. |
|---|---|
| Journal, issue, pages | Nature, Vol. 584, Issue 7819, Page 125-129, Year 2020 |
| Publish date | Jun 11, 2020 |
 Authors Authors | Jie Yin / Kuang-Yui M Chen / Mary J Clark / Mahdi Hijazi / Punita Kumari / Xiao-Chen Bai / Roger K Sunahara / Patrick Barth / Daniel M Rosenbaum /   |
| PubMed Abstract | The D2 dopamine receptor (DRD2) is a therapeutic target for Parkinson's disease and antipsychotic drugs. DRD2 is activated by the endogenous neurotransmitter dopamine and synthetic agonist drugs such ...The D2 dopamine receptor (DRD2) is a therapeutic target for Parkinson's disease and antipsychotic drugs. DRD2 is activated by the endogenous neurotransmitter dopamine and synthetic agonist drugs such as bromocriptine, leading to stimulation of G and inhibition of adenylyl cyclase. Here we used cryo-electron microscopy to elucidate the structure of an agonist-bound activated DRD2-G complex reconstituted into a phospholipid membrane. The extracellular ligand-binding site of DRD2 is remodelled in response to agonist binding, with conformational changes in extracellular loop 2, transmembrane domain 5 (TM5), TM6 and TM7, propagating to opening of the intracellular G-binding site. The DRD2-G structure represents, to our knowledge, the first experimental model of a G-protein-coupled receptor-G-protein complex embedded in a phospholipid bilayer, which serves as a benchmark to validate the interactions seen in previous detergent-bound structures. The structure also reveals interactions that are unique to the membrane-embedded complex, including helix 8 burial in the inner leaflet, ordered lysine and arginine side chains in the membrane interfacial regions, and lipid anchoring of the G protein in the membrane. Our model of the activated DRD2 will help to inform the design of subtype-selective DRD2 ligands for multiple human central nervous system disorders. |
 External links External links |  Nature / Nature /  PubMed:32528175 / PubMed:32528175 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 3.7 - 3.8 Å |
| Structure data | EMDB-21243, PDB-6vms:  EMDB-21244:  EMDB-21245: |
| Chemicals | 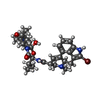 ChemComp-08Y: |
| Source |
|
 Keywords Keywords | SIGNALING PROTEIN / Dopamine / Dopamine receptor / GPCR / G protein / Parkinson's disease |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers





 homo sapiens (human)
homo sapiens (human)
 enterobacteria phage t4 (virus)
enterobacteria phage t4 (virus)