+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21151 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of HIV-1 conserved Intasome Core | |||||||||
 Map data Map data | HIV-1 conserved Intasome Core | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | site-specific recombination / retroviruses / integrase / integration / nucleoprotein complex / DNA complex / integrase strand transfer inhibitor / TRANSFERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / endonuclease activity / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / host cell cytoplasm / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Li M / Chen X | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies. Authors: Min Li / Xuemin Chen / Huaibin Wang / Kellie A Jurado / Alan N Engelman / Robert Craigie /  Abstract: The low solubility and aggregation properties of HIV-1 integrase (IN) are major obstacles for biochemical and structural studies. The lens epithelium-derived growth factor (LEDGF) is a cellular ...The low solubility and aggregation properties of HIV-1 integrase (IN) are major obstacles for biochemical and structural studies. The lens epithelium-derived growth factor (LEDGF) is a cellular factor that binds IN and tethers preintegration complexes to chromatin before integration. The LEDGF also stimulates HIV-1 IN DNA strand transfer activity and improves its solubility in vitro. We show that these properties are conferred by a short peptide spanning residues 178 to 197 of the LEDGF that encompasses its AT-hook DNA-binding elements. The peptide stimulates HIV-1 IN activity both in trans and in cis. Fusion of the peptide to either the N- or C-terminus of IN results in maximal stimulation of concerted integration activity and greatly improves the solubility of the protein and nucleoprotein complexes of IN with viral DNA ends (intasomes). High-resolution structures of HIV-1 intasomes are required to understand the mechanism of IN strand transfer inhibitors (INSTIs), which are front-line drugs for the treatment of HIV-1, and how the virus can develop resistance to INSTIs. We have previously determined the structure of the HIV-1 strand transfer complex intasome. The improved biophysical properties of intasomes assembled with LEDGF peptide fusion IN have enabled us to determine the structure of the cleaved synaptic complex intasome, which is the direct target of INSTIs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21151.map.gz emd_21151.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21151-v30.xml emd-21151-v30.xml emd-21151.xml emd-21151.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21151.png emd_21151.png | 70.4 KB | ||
| Filedesc metadata |  emd-21151.cif.gz emd-21151.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21151 http://ftp.pdbj.org/pub/emdb/structures/EMD-21151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21151 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21151 | HTTPS FTP |
-Related structure data
| Related structure data |  6vdkMC  6u8qC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21151.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21151.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV-1 conserved Intasome Core | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 conserved intasome core
| Entire | Name: HIV-1 conserved intasome core |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 conserved intasome core
| Supramolecule | Name: HIV-1 conserved intasome core / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: Integrase
| Macromolecule | Name: Integrase / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO EC number: Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 39.898355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMPKRGRP AATEVKIPKP RGRPPLPAGT NSKGPPDFSS DEEREPTPVL GSGAAAAGQS RAAVGRKATK KTDGGGFLDG IDKAQEEHE KYHSNWRAMA SDFNLPPVVA KEIVASCDKC QLKGEAMHGQ VDCSPGIWQL DCTHLEGKVI LVAVHVASGY I EAEVIPAE ...String: GSHMPKRGRP AATEVKIPKP RGRPPLPAGT NSKGPPDFSS DEEREPTPVL GSGAAAAGQS RAAVGRKATK KTDGGGFLDG IDKAQEEHE KYHSNWRAMA SDFNLPPVVA KEIVASCDKC QLKGEAMHGQ VDCSPGIWQL DCTHLEGKVI LVAVHVASGY I EAEVIPAE TGQETAYFLL KLAGRWPVKT VHTDNGSNFT STTVKAACWW AGIKQEFGIP YNPQSQGVIE SMNKELKKII GQ VRDQAEH LKTAVQMAVF IHNFKRKGGI GGYSAGERIV DIIATDIQTK ELQKQITKIQ NFRVYYRDSR DPVWKGPAKL LWK GEGAVV IQDNSDIKVV PRRKAKIIRD YGKQMAGDDC VASRQDED UniProtKB: Gag-Pol polyprotein |
-Macromolecule #2: DNA (27-MER)
| Macromolecule | Name: DNA (27-MER) / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.188271 KDa |
| Sequence | String: (DA)(DC)(DT)(DG)(DC)(DT)(DA)(DG)(DA)(DG) (DA)(DT)(DT)(DT)(DT)(DC)(DC)(DC)(DG)(DC) (DC)(DC)(DA)(DC)(DG)(DC)(DT) |
-Macromolecule #3: DNA (25-MER)
| Macromolecule | Name: DNA (25-MER) / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.773023 KDa |
| Sequence | String: (DA)(DG)(DC)(DG)(DT)(DG)(DG)(DG)(DC)(DG) (DG)(DG)(DA)(DA)(DA)(DA)(DT)(DC)(DT)(DC) (DT)(DA)(DG)(DC)(DA) |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4...
| Macromolecule | Name: (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide type: ligand / ID: 5 / Number of copies: 2 / Formula: DLU |
|---|---|
| Molecular weight | Theoretical: 419.379 Da |
| Chemical component information | 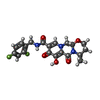 ChemComp-DLU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















