+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20772 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fab397 in complex with NPNA8 peptide (Class 1) | |||||||||
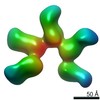
 Map data Map data | Fab397 in complex with NPNA8 peptide (Class 1) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
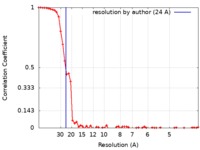
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Ward AB / Torres JL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2020 Journal: J Mol Biol / Year: 2020Title: Diverse Antibody Responses to Conserved Structural Motifs in Plasmodium falciparum Circumsporozoite Protein. Authors: Tossapol Pholcharee / David Oyen / Jonathan L Torres / Yevel Flores-Garcia / Gregory M Martin / Gonzalo E González-Páez / Daniel Emerling / Wayne Volkmuth / Emily Locke / C Richter King / ...Authors: Tossapol Pholcharee / David Oyen / Jonathan L Torres / Yevel Flores-Garcia / Gregory M Martin / Gonzalo E González-Páez / Daniel Emerling / Wayne Volkmuth / Emily Locke / C Richter King / Fidel Zavala / Andrew B Ward / Ian A Wilson /  Abstract: Malaria vaccine candidate RTS,S/AS01 is based on the central and C-terminal regions of the circumsporozoite protein (CSP) of P. falciparum. mAb397 was isolated from a volunteer in an RTS,S/AS01 ...Malaria vaccine candidate RTS,S/AS01 is based on the central and C-terminal regions of the circumsporozoite protein (CSP) of P. falciparum. mAb397 was isolated from a volunteer in an RTS,S/AS01 clinical trial, and it protects mice from infection by malaria sporozoites. However, mAb397 originates from the less commonly used VH3-15 germline gene compared to the VH3-30/33 antibodies generally elicited by RTS,S to the central NANP repeat region of CSP. The crystal structure of mAb397 with an NPNA peptide shows that the central NPNA forms a type I β-turn and is the main recognition motif. In most anti-NANP antibodies studied to date, a germline-encoded Trp is used to engage the Pro in NPNA β-turns, but here the Trp interacts with the first Asn. This "conserved" Trp, however, can arise from different germline genes and be located in the heavy or the light chain. Variation in the terminal ψ angles of the NPNA β-turns results in different dispositions of the subsequent NPNA and, hence, different stoichiometries and modes of antibody binding to rsCSP. Diverse protective antibodies against NANP repeats are therefore not limited to a single germline gene response or mode of binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20772.map.gz emd_20772.map.gz | 14.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20772-v30.xml emd-20772-v30.xml emd-20772.xml emd-20772.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20772_fsc.xml emd_20772_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_20772.png emd_20772.png | 35.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20772 http://ftp.pdbj.org/pub/emdb/structures/EMD-20772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20772 | HTTPS FTP |
-Validation report
| Summary document |  emd_20772_validation.pdf.gz emd_20772_validation.pdf.gz | 77.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20772_full_validation.pdf.gz emd_20772_full_validation.pdf.gz | 76.6 KB | Display | |
| Data in XML |  emd_20772_validation.xml.gz emd_20772_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20772 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20772 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20772 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20772 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20772.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20772.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Fab397 in complex with NPNA8 peptide (Class 1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Fragment antigen binding (Fab) 397 in complex with NPNA8 peptide ...
| Entire | Name: Fragment antigen binding (Fab) 397 in complex with NPNA8 peptide (Class 1) |
|---|---|
| Components |
|
-Supramolecule #1: Fragment antigen binding (Fab) 397 in complex with NPNA8 peptide ...
| Supramolecule | Name: Fragment antigen binding (Fab) 397 in complex with NPNA8 peptide (Class 1) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 150 KDa |
-Supramolecule #2: fragment antigen binding (fab) 397
| Supramolecule | Name: fragment antigen binding (fab) 397 / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: NPNA8 Peptide
| Supramolecule | Name: NPNA8 Peptide / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: 2% Uranyl Formate | |||||||||
| Grid | Model: Homemade / Material: COPPER / Support film - Material: CELLULOSE ACETATE / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.2 mm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera


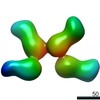

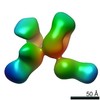
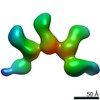










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)