+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20612 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
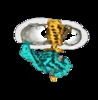
| Title | GPCR-Beta arrestin structure in lipid bilayer | |||||||||
 Map data Map data | GPCR-Beta arrestin structure in lipid bilayer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Arrestin / GPCR / complex / signaling / SIGNALING PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationV2 vasopressin receptor binding / alpha-1A adrenergic receptor binding / follicle-stimulating hormone receptor binding / TGFBR3 regulates TGF-beta signaling / sensory perception of touch / G alpha (s) signalling events / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / follicle-stimulating hormone signaling pathway / regulation of inositol trisphosphate biosynthetic process ...V2 vasopressin receptor binding / alpha-1A adrenergic receptor binding / follicle-stimulating hormone receptor binding / TGFBR3 regulates TGF-beta signaling / sensory perception of touch / G alpha (s) signalling events / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / follicle-stimulating hormone signaling pathway / regulation of inositol trisphosphate biosynthetic process / protein phosphorylated amino acid binding / alpha-1B adrenergic receptor binding / Vasopressin-like receptors / regulation of systemic arterial blood pressure by vasopressin / vasopressin receptor activity / Lysosome Vesicle Biogenesis / phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway / AP-2 adaptor complex binding / Ub-specific processing proteases / angiotensin receptor binding / Muscarinic acetylcholine receptors / cholinergic synapse / G protein-coupled acetylcholine receptor activity / MAP2K and MAPK activation / Golgi Associated Vesicle Biogenesis / hemostasis / Cargo recognition for clathrin-mediated endocytosis / clathrin-cargo adaptor activity / telencephalon development / Clathrin-mediated endocytosis / regulation of smooth muscle contraction / negative regulation of interleukin-8 production / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / regulation of G protein-coupled receptor signaling pathway / cysteine-type endopeptidase inhibitor activity involved in apoptotic process / arrestin family protein binding / G protein-coupled receptor internalization / mitogen-activated protein kinase kinase binding / Thrombin signalling through proteinase activated receptors (PARs) / clathrin binding / response to morphine / stress fiber assembly / positive regulation of Rho protein signal transduction / regulation of heart contraction / pseudopodium / positive regulation of systemic arterial blood pressure / negative regulation of interleukin-6 production / positive regulation of intracellular signal transduction / positive regulation of receptor internalization / positive regulation of vasoconstriction / negative regulation of Notch signaling pathway / phototransduction / endocytic vesicle / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / immunoglobulin complex / positive regulation of insulin secretion involved in cellular response to glucose stimulus / activation of adenylate cyclase activity / cellular response to hormone stimulus / insulin-like growth factor receptor binding / response to cytokine / clathrin-coated pit / negative regulation of protein ubiquitination / presynaptic modulation of chemical synaptic transmission / GTPase activator activity / positive regulation of protein ubiquitination / nuclear estrogen receptor binding / phosphoprotein binding / clathrin-coated endocytic vesicle membrane / G protein-coupled receptor binding / negative regulation of ERK1 and ERK2 cascade / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to virus / endocytosis / positive regulation of protein phosphorylation / G protein-coupled acetylcholine receptor signaling pathway / Vasopressin regulates renal water homeostasis via Aquaporins / nervous system development / Cargo recognition for clathrin-mediated endocytosis / protein transport / presynapse / Clathrin-mediated endocytosis / cytoplasmic vesicle / regulation of apoptotic process / G alpha (i) signalling events / G alpha (s) signalling events / basolateral plasma membrane / dendritic spine / ubiquitin-dependent protein catabolic process / chemical synaptic transmission / negative regulation of neuron apoptotic process / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / adaptive immune response / transmembrane transporter binding / postsynaptic membrane / transcription coactivator activity / positive regulation of ERK1 and ERK2 cascade / positive regulation of MAPK cascade / endosome / postsynaptic density Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Staus DP / Hu H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure of the M2 muscarinic receptor-β-arrestin complex in a lipid nanodisc. Authors: Dean P Staus / Hongli Hu / Michael J Robertson / Alissa L W Kleinhenz / Laura M Wingler / William D Capel / Naomi R Latorraca / Robert J Lefkowitz / Georgios Skiniotis /   Abstract: After activation by an agonist, G-protein-coupled receptors (GPCRs) recruit β-arrestin, which desensitizes heterotrimeric G-protein signalling and promotes receptor endocytosis. Additionally, β- ...After activation by an agonist, G-protein-coupled receptors (GPCRs) recruit β-arrestin, which desensitizes heterotrimeric G-protein signalling and promotes receptor endocytosis. Additionally, β-arrestin directly regulates many cell signalling pathways that can induce cellular responses distinct from that of G proteins. In contrast to G proteins, for which there are many high-resolution structures in complex with GPCRs, the molecular mechanisms underlying the interaction of β-arrestin with GPCRs are much less understood. Here we present a cryo-electron microscopy structure of β-arrestin 1 (βarr1) in complex with M2 muscarinic receptor (M2R) reconstituted in lipid nanodiscs. The M2R-βarr1 complex displays a multimodal network of flexible interactions, including binding of the N domain of βarr1 to phosphorylated receptor residues and insertion of the finger loop of βarr1 into the M2R seven-transmembrane bundle, which adopts a conformation similar to that in the M2R-heterotrimeric G protein complex. Moreover, the cryo-electron microscopy map reveals that the C-edge of βarr1 engages the lipid bilayer. Through atomistic simulations and biophysical, biochemical and cellular assays, we show that the C-edge is critical for stable complex formation, βarr1 recruitment, receptor internalization, and desensitization of G-protein activation. Taken together, these data suggest that the cooperative interactions of β-arrestin with both the receptor and the phospholipid bilayer contribute to its functional versatility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20612.map.gz emd_20612.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20612-v30.xml emd-20612-v30.xml emd-20612.xml emd-20612.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20612.png emd_20612.png | 96.1 KB | ||
| Filedesc metadata |  emd-20612.cif.gz emd-20612.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20612 http://ftp.pdbj.org/pub/emdb/structures/EMD-20612 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20612 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20612 | HTTPS FTP |
-Related structure data
| Related structure data |  6u1nMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20612.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20612.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GPCR-Beta arrestin structure in lipid bilayer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Phosphorylated human muscarinic acetylcholine receptor M2 in comp...
| Entire | Name: Phosphorylated human muscarinic acetylcholine receptor M2 in complex with rat beta-arrestin1, stabilized by an antibody fragment (Fab30). |
|---|---|
| Components |
|
-Supramolecule #1: Phosphorylated human muscarinic acetylcholine receptor M2 in comp...
| Supramolecule | Name: Phosphorylated human muscarinic acetylcholine receptor M2 in complex with rat beta-arrestin1, stabilized by an antibody fragment (Fab30). type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: Phosphorylated human muscarinic acetylcholine receptor M2
| Supramolecule | Name: Phosphorylated human muscarinic acetylcholine receptor M2 type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Phosphorylated M2R was generated by ligating a synthetic phosphopeptide derived from the vasopressin-2-receptor (V2Rpp) using the enzyme sortase. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Cysteine-free rat beta-arrestin 1 truncated at amino acid 393
| Supramolecule | Name: Cysteine-free rat beta-arrestin 1 truncated at amino acid 393 type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: Antibody Fragment (Fab30)
| Supramolecule | Name: Antibody Fragment (Fab30) / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Muscarinic acetylcholine receptor M2, Vasopressin V2 receptor chimera
| Macromolecule | Name: Muscarinic acetylcholine receptor M2, Vasopressin V2 receptor chimera type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.616176 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKNN STNSSNNSLA LTSPYKTFEV VFIVLVAGSL SLVTIIGNIL VMVSIKVNRH LQTVNNYFLF SLACADLIIG VFSMNLYTL YTVIGYWPLG PVVCDLWLAL DYVVSNASVM NLLIISFDRY FCVTKPLTYP VKRTTKMAGM MIAAAWVLSF I LWAPAILF ...String: DYKDDDDKNN STNSSNNSLA LTSPYKTFEV VFIVLVAGSL SLVTIIGNIL VMVSIKVNRH LQTVNNYFLF SLACADLIIG VFSMNLYTL YTVIGYWPLG PVVCDLWLAL DYVVSNASVM NLLIISFDRY FCVTKPLTYP VKRTTKMAGM MIAAAWVLSF I LWAPAILF WQFIVGVRTV EDGECYIQFF SNAAVTFGTA IAAFYLPVII MTVLYWHISR ASKSRIKKDK KEPVANQDPV SP SLVQGRI VKPNNNNMPS SDDGLEHNKI QNGKAPRDPV TENCVQGEEK ESSNDSTSVS AVASNMRDDE ITQDENTVST SLG HSKDEN SKQTCIRIGT KTPKSDSCTP TNTTVEVVGS SGQNGDEKQN IVARKIVKMT KQPAKKKPPP SREKKVTRTI LAIL LAFII TWAPYNVMVL INTFCAPCIP NTVWTIGYWL CYINSTINPA CYALCNATFK KTFKHLLMCH YKNIGATRLP ETGGG ARGR TPPSLGPQDE (SEP)C(TPO)(TPO)A(SEP)(SEP)(SEP)LA KDTSS UniProtKB: Muscarinic acetylcholine receptor M2, Vasopressin V2 receptor |
-Macromolecule #2: Beta-arrestin-1
| Macromolecule | Name: Beta-arrestin-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.01718 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSPEFPGRLG DKGTRVFKKA SPNGKLTVYL GKRDFVDHID LVDPVDGVVL VDPEYLKERR VYVTLTVAFR YGREDLDVLG LTFRKDLFV ANVQSFPPAP EDKKPLTRLQ ERLIKKLGEH AYPFTFEIPP NLPSSVTLQP GPEDTGKALG VDYEVKAFVA E NLEEKIHK ...String: GSPEFPGRLG DKGTRVFKKA SPNGKLTVYL GKRDFVDHID LVDPVDGVVL VDPEYLKERR VYVTLTVAFR YGREDLDVLG LTFRKDLFV ANVQSFPPAP EDKKPLTRLQ ERLIKKLGEH AYPFTFEIPP NLPSSVTLQP GPEDTGKALG VDYEVKAFVA E NLEEKIHK RNSVRLVIRK VQYAPERPGP QPTAETTRQF LMSDKPLHLE ASLDKEIYYH GEPISVNVHV TNNTNKTVKK IK ISVRQYA DIVLFNTAQY KVPVAMEEAD DTVAPSSTFS KVYTLTPFLA NNREKRGLAL DGKLKHEDTN LASSTLLREG ANR EILGII VSYKVKVKLV VSRGGLLGDL ASSDVAVELP FTLMHPKPKE EPPHREVPES ETPVDTNLIE LDTNDDDIVF EDFA R UniProtKB: Beta-arrestin-1 |
-Macromolecule #3: Fab30 heavy chain
| Macromolecule | Name: Fab30 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.512354 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYYGY TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSRQFWYSG LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYYGY TYYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARSRQFWYSG LDYWGQGTLV TVSSASTKGP SVFPLAPSSK STSGGTAALG CLVKDYFPEP V TVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KKVEPKSCDK THHHHHHHH UniProtKB: Epididymis luminal protein 214 |
-Macromolecule #4: Fab30 light chain
| Macromolecule | Name: Fab30 light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.435064 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYKYVPVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQYKYVPVTF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC UniProtKB: Ig-like domain-containing protein |
-Macromolecule #5: 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1...
| Macromolecule | Name: 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide type: ligand / ID: 5 / Number of copies: 1 / Formula: 2CU |
|---|---|
| Molecular weight | Theoretical: 437.944 Da |
| Chemical component information |  ChemComp-2CU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 47169 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 47169 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6u1n: |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















