[English] 日本語
 Yorodumi
Yorodumi- EMDB-20114: CryoEM structure of PilB from Geobacter metallireducens: C2ccocco... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20114 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of PilB from Geobacter metallireducens: C2ccocco conformation | |||||||||
 Map data Map data | Sharpened and z-flipped map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATPase / T4P / type iv pilus / motor / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpilus assembly / ATP hydrolysis activity / ATP binding / metal ion binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Geobacter metallireducens (bacteria) / Geobacter metallireducens (bacteria) /  Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria) Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria) | |||||||||
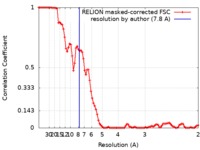
| Method | single particle reconstruction / cryo EM / Resolution: 7.8 Å | |||||||||
 Authors Authors | McCallum M / Howell PL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Multiple conformations facilitate PilT function in the type IV pilus. Authors: Matthew McCallum / Samir Benlekbir / Sheryl Nguyen / Stephanie Tammam / John L Rubinstein / Lori L Burrows / P Lynne Howell /  Abstract: Type IV pilus-like systems are protein complexes that polymerize pilin fibres. They are critical for virulence in many bacterial pathogens. Pilin polymerization and depolymerization are powered by ...Type IV pilus-like systems are protein complexes that polymerize pilin fibres. They are critical for virulence in many bacterial pathogens. Pilin polymerization and depolymerization are powered by motor ATPases of the PilT/VirB11-like family. This family is thought to operate with C symmetry; however, most of these ATPases crystallize with either C or C symmetric conformations. The relevance of these conformations is unclear. Here, we determine the X-ray structures of PilT in four unique conformations and use these structures to classify the conformation of available PilT/VirB11-like family member structures. Single particle electron cryomicroscopy (cryoEM) structures of PilT reveal condition-dependent preferences for C C, and C conformations. The physiologic importance of these conformations is validated by coevolution analysis and functional studies of point mutants, identifying a rare gain-of-function mutation that favours the C conformation. With these data, we propose a comprehensive model of PilT function with broad implications for PilT/VirB11-like family members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20114.map.gz emd_20114.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20114-v30.xml emd-20114-v30.xml emd-20114.xml emd-20114.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20114_fsc.xml emd_20114_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20114.png emd_20114.png | 91.6 KB | ||
| Others |  emd_20114_additional.map.gz emd_20114_additional.map.gz | 31.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20114 http://ftp.pdbj.org/pub/emdb/structures/EMD-20114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20114 | HTTPS FTP |
-Related structure data
| Related structure data |  6oljMC  6ojxC  6ojyC  6ojzC  6ok2C  6okvC  6olkC  6ollC  6olmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20114.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20114.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and z-flipped map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Original unsharpened and unflipped map
| File | emd_20114_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Original unsharpened and unflipped map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PilB Hexamer
| Entire | Name: PilB Hexamer |
|---|---|
| Components |
|
-Supramolecule #1: PilB Hexamer
| Supramolecule | Name: PilB Hexamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Geobacter metallireducens (bacteria) Geobacter metallireducens (bacteria) |
-Macromolecule #1: Type IV pilus biogenesis ATPase PilB
| Macromolecule | Name: Type IV pilus biogenesis ATPase PilB / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria) Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria)Strain: GS-15 / ATCC 53774 / DSM 7210 |
| Molecular weight | Theoretical: 64.899281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MQASRLGELL VRNNVITKEQ LAKALEEQKS ADGQQRLGSI LIKNGLISEP DLTSFLSKQY GVPSINLSE FEAEQAVVKI IPADVAQKYQ IVPVNRAGST LIIAMADPSN IFAIDDIKFM TGYNVEVVVA SESAIKAAID K YYDQSASL ...String: MGSSHHHHHH SSGLVPRGSH MQASRLGELL VRNNVITKEQ LAKALEEQKS ADGQQRLGSI LIKNGLISEP DLTSFLSKQY GVPSINLSE FEAEQAVVKI IPADVAQKYQ IVPVNRAGST LIIAMADPSN IFAIDDIKFM TGYNVEVVVA SESAIKAAID K YYDQSASL ADVMGDLEMD DLEVIDTDDE VDVSSLERAT EDAPVVKLVN LILTDAIKRK ASDIHIEPYE RSFRVRYRID GV LYEVMKP PLKLKNAITS RIKIMAELDI AERRLPQDGR IKIKLGGGQD MDYRVSVLPT LFGEKVVLRL LDKSNLQLDM TKL GYEPDA LHYFKEAIHK PFGMVLVTGP TGSGKTVSLY SALGELNKTT ENISTAEDPV EFNFAGINQV QMHEDIGLNF AAAL RSFLR QDPDIIMIGE IRDFETAEIA IKAALTGHLV LSTLHTNDAP ATINRLLNMG VEPFLVASAV NLITAQRLAR RVCSE CKQP EEIPIQALID AGVSPDEAPS YVCYKGTGCV KCNNTGYKGR VGFYQVMPML EEIRELILNG ANTAEIKRES MRLGIK TMR QSGLTKLKEG VTSFEEVLRV TVADD UniProtKB: Type IV pilus biogenesis ATPase PilB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Homemade / Material: GOLD |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 35.7 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)