[English] 日本語
 Yorodumi
Yorodumi- EMDB-20116: CryoEM structure of PilT4 from Geobacter metallireducens without ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20116 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
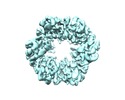
| Title | CryoEM structure of PilT4 from Geobacter metallireducens without adding nucleotide: C2oocooc conformation | |||||||||
 Map data Map data | Sharpened and z-flipped map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATPase / T4P / type iv pilus / motor / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Geobacter metallireducens (bacteria) / Geobacter metallireducens (bacteria) /  Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria) Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria) | |||||||||
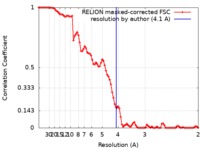
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | McCallum M / Howell PL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Multiple conformations facilitate PilT function in the type IV pilus. Authors: Matthew McCallum / Samir Benlekbir / Sheryl Nguyen / Stephanie Tammam / John L Rubinstein / Lori L Burrows / P Lynne Howell /  Abstract: Type IV pilus-like systems are protein complexes that polymerize pilin fibres. They are critical for virulence in many bacterial pathogens. Pilin polymerization and depolymerization are powered by ...Type IV pilus-like systems are protein complexes that polymerize pilin fibres. They are critical for virulence in many bacterial pathogens. Pilin polymerization and depolymerization are powered by motor ATPases of the PilT/VirB11-like family. This family is thought to operate with C symmetry; however, most of these ATPases crystallize with either C or C symmetric conformations. The relevance of these conformations is unclear. Here, we determine the X-ray structures of PilT in four unique conformations and use these structures to classify the conformation of available PilT/VirB11-like family member structures. Single particle electron cryomicroscopy (cryoEM) structures of PilT reveal condition-dependent preferences for C C, and C conformations. The physiologic importance of these conformations is validated by coevolution analysis and functional studies of point mutants, identifying a rare gain-of-function mutation that favours the C conformation. With these data, we propose a comprehensive model of PilT function with broad implications for PilT/VirB11-like family members. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20116.map.gz emd_20116.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20116-v30.xml emd-20116-v30.xml emd-20116.xml emd-20116.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20116_fsc.xml emd_20116_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20116.png emd_20116.png | 126.8 KB | ||
| Filedesc metadata |  emd-20116.cif.gz emd-20116.cif.gz | 5.5 KB | ||
| Others |  emd_20116_additional.map.gz emd_20116_additional.map.gz | 31.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20116 http://ftp.pdbj.org/pub/emdb/structures/EMD-20116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20116 | HTTPS FTP |
-Related structure data
| Related structure data |  6ollMC  6ojxC  6ojyC  6ojzC  6ok2C  6okvC  6oljC  6olkC  6olmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20116.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20116.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and z-flipped map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Original unsharpened and unflipped map
| File | emd_20116_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Original unsharpened and unflipped map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PilT Hexamer
| Entire | Name: PilT Hexamer |
|---|---|
| Components |
|
-Supramolecule #1: PilT Hexamer
| Supramolecule | Name: PilT Hexamer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Geobacter metallireducens (bacteria) Geobacter metallireducens (bacteria) |
-Macromolecule #1: Twitching motility pilus retraction ATPase
| Macromolecule | Name: Twitching motility pilus retraction ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria) Geobacter metallireducens (strain GS-15 / ATCC 53774 / DSM 7210) (bacteria)Strain: GS-15 / ATCC 53774 / DSM 7210 |
| Molecular weight | Theoretical: 42.883379 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MANMHQLLTE LVNRGGSDLH LTTNSPPQIR IDGKLLPLDM PPLNAVDTKQ LCYSILTEQQ KHKFEENNE LDLSFGIKGL SRFRGNVFVQ RGAVAGVFRV IPYKILSFEE LGLPPVVREL AEKPRGLVLV TGPTGSGKST T LAAIIDKI ...String: MGSSHHHHHH SSGLVPRGSH MANMHQLLTE LVNRGGSDLH LTTNSPPQIR IDGKLLPLDM PPLNAVDTKQ LCYSILTEQQ KHKFEENNE LDLSFGIKGL SRFRGNVFVQ RGAVAGVFRV IPYKILSFEE LGLPPVVREL AEKPRGLVLV TGPTGSGKST T LAAIIDKI NTDRHEHIVT VEDPIEYLHP HKSCVVNQRE VGADTKSFKN ALKYILRQDP DVVLVGELRD LETIEAALTL AE TGHLCFA TLHTNSAVQT INRIVDVFPS YQQPQVRAQL SFVLEGVLSQ TLLPKASGTG RVLAIEVMVP NPAIRNLIRE DKI HQIYSQ MQVGQEKFGM MTMNQCLYGL LQKRHITMDV GMGRSPDPDE LKQMLTSGVR PQAPRPPMR UniProtKB: Twitching motility pilus retraction ATPase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Homemade / Material: GOLD |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 42.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial fitting in chimera, flexible fitting in Phenix-refine |
| Refinement | Protocol: FLEXIBLE FIT / Overall B value: 38.4 |
| Output model |  PDB-6oll: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)