+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20053 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of YenTcA in its prepore state | |||||||||||||||
 Map data Map data | em-volume_P1 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Membrane protein Pore-forming toxin Complex / TOXIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationendochitinase activity / chitinase / chitin catabolic process / chitin binding / polysaccharide catabolic process / extracellular region Similarity search - Function | |||||||||||||||
| Biological species |  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) | |||||||||||||||
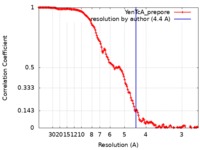
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||||||||
 Authors Authors | Piper SJ / Brillault L | |||||||||||||||
| Funding support |  Australia, Australia,  New Zealand, 4 items New Zealand, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin. Authors: Sarah J Piper / Lou Brillault / Rosalba Rothnagel / Tristan I Croll / Joseph K Box / Irene Chassagnon / Sebastian Scherer / Kenneth N Goldie / Sandra A Jones / Femke Schepers / Lauren ...Authors: Sarah J Piper / Lou Brillault / Rosalba Rothnagel / Tristan I Croll / Joseph K Box / Irene Chassagnon / Sebastian Scherer / Kenneth N Goldie / Sandra A Jones / Femke Schepers / Lauren Hartley-Tassell / Thomas Ve / Jason N Busby / Julie E Dalziel / J Shaun Lott / Ben Hankamer / Henning Stahlberg / Mark R H Hurst / Michael J Landsberg /      Abstract: ABC toxins are pore-forming virulence factors produced by pathogenic bacteria. YenTcA is the pore-forming and membrane binding A subunit of the ABC toxin YenTc, produced by the insect pathogen ...ABC toxins are pore-forming virulence factors produced by pathogenic bacteria. YenTcA is the pore-forming and membrane binding A subunit of the ABC toxin YenTc, produced by the insect pathogen Yersinia entomophaga. Here we present cryo-EM structures of YenTcA, purified from the native source. The soluble pre-pore structure, determined at an average resolution of 4.4 Å, reveals a pentameric assembly that in contrast to other characterised ABC toxins is formed by two TcA-like proteins (YenA1 and YenA2) and decorated by two endochitinases (Chi1 and Chi2). We also identify conformational changes that accompany membrane pore formation by visualising YenTcA inserted into liposomes. A clear outward rotation of the Chi1 subunits allows for access of the protruding translocation pore to the membrane. Our results highlight structural and functional diversity within the ABC toxin subfamily, explaining how different ABC toxins are capable of recognising diverse hosts. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20053.map.gz emd_20053.map.gz | 16.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20053-v30.xml emd-20053-v30.xml emd-20053.xml emd-20053.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20053_fsc.xml emd_20053_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_20053.png emd_20053.png | 76.8 KB | ||
| Filedesc metadata |  emd-20053.cif.gz emd-20053.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20053 http://ftp.pdbj.org/pub/emdb/structures/EMD-20053 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20053 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20053 | HTTPS FTP |
-Related structure data
| Related structure data |  6ogdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20053.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20053.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-volume_P1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yersinia entomophaga toxin complex subunit A (YenTcA) in the pre-...
| Entire | Name: Yersinia entomophaga toxin complex subunit A (YenTcA) in the pre-pore form |
|---|---|
| Components |
|
-Supramolecule #1: Yersinia entomophaga toxin complex subunit A (YenTcA) in the pre-...
| Supramolecule | Name: Yersinia entomophaga toxin complex subunit A (YenTcA) in the pre-pore form type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) / Strain: MH96 Yersinia entomophaga (bacteria) / Strain: MH96 |
| Molecular weight | Theoretical: 2.08 MDa |
-Macromolecule #1: Toxin subunit YenA1
| Macromolecule | Name: Toxin subunit YenA1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Molecular weight | Theoretical: 129.91232 KDa |
| Sequence | String: MDKYNNYSNV IKNKSSISPL LAAAAKIEPE ITVLSSASKS NRSQYSQSLA DTLLGLGYRS IFDIAKVSRQ RFIKRHDESL LGNGAVIFD KAVSMANQVL QKYRKNRLEK SNSPLVPQTS SSTDASSESQ TNKLPEYNQL FPEPWDNFCR PGAIEALDSP A SYLLDLYK ...String: MDKYNNYSNV IKNKSSISPL LAAAAKIEPE ITVLSSASKS NRSQYSQSLA DTLLGLGYRS IFDIAKVSRQ RFIKRHDESL LGNGAVIFD KAVSMANQVL QKYRKNRLEK SNSPLVPQTS SSTDASSESQ TNKLPEYNQL FPEPWDNFCR PGAIEALDSP A SYLLDLYK FIQSVELDGS NQARKLETRR ADIPKLSLDN DALYKEVTAL SIVNDVLSGS AREYIDQSGQ ADKAVNQILG DT HFPFTLP YSLPTQQINK GLGASNIELG TVIQRVDPQF SWNTTQEKYN QVLLAYTQLS SEQIALLSLP DVFTQNFLTQ TEL SAGYLS ASTTEILAEK DLSRHGYIVK AADNIKGPTQ LVEHSDASYD VIELTCTNQA KETITVKLRG ENIITYQRTK ARMV PFDNS SPFSRQLKLT FVAEDNPSLG NLDKGPYFAN MDIYAAEWVR ENVSSETMVS RPFLTMTYRI AIAKAGASLE ELQPE ADAF FINNFGLSAE DSSQLVKLVA FGDQTGSKAE EIESLLSCGE NLPIVSPNVI FANPIFGSYF NDEPFPAPYH FGGVYI NAH QRNAMTIIRA EGGREIQSLS NFRLERLNRF IRLQRWLDLP SHQLDLLLTS VMQADADNSQ QEITEPVLKS LGLFRHL NL QYKITPEIFS SWLYQLTPFA VSGEIAFFDR IFNREQLFDQ PFILDGGSFT YLDAKGSDAK SVKQLCAGLN ISAVTFQF I APLVQSALGL EAGTLVRSFE VVSSLYRLVS IPQTFGLSTE DGLILMNILT DEMGYLAKQP AFDDKQTQDK DFLSIILKM EALSAWLTKN NLTPASLALL LGVTRLAVVP TNNMVTFFKG IANGLSENVC LTTDDFQRQE LEGADWWTLL STNQVIDDMG LVLDIHPVW GKSDEEMLME KIQSIGVSND NNTLSIIVQI LIQAKNAQEN LLSQTISAEY GVERSVVPLQ LRWLGSNVYS V LNQVLNNT PTDISSIVPK LSELTYSLLI YTQLINSLKL NKEFIFLRLT QPNWLGLTQP KLSTQLSLPE IYLITCYQDW VV NANKNED SIHEYLEFAN IKKTEAEKTL VDNSEKCAEL LAEILAWDAG EILKAASLLG LNPPQATNVF EIDWIRRLQT LSE KTMIST EYLWQMGDLT ENSEFSLKEG VGEAVMAALK AQGDSDNV UniProtKB: Toxin subunit YenA1 |
-Macromolecule #2: Toxin subunit YenA2
| Macromolecule | Name: Toxin subunit YenA2 / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Molecular weight | Theoretical: 156.324938 KDa |
| Sequence | String: MSNSIEAKLQ EDLRDALVDY YLGQIVPNSK DFTNLRSTIK NVDDLYDHLL LDTQVSAKVI TSRLSLVTQS VQQYINRIAL NLEPGLSIN QQEATDWEEF ANRYGYWAAN QQLRMFPEIY VDPTLRLTKT EFFFQLESAL NQGKLTDDVA QKAVLGYLNN F EEVSNLEI ...String: MSNSIEAKLQ EDLRDALVDY YLGQIVPNSK DFTNLRSTIK NVDDLYDHLL LDTQVSAKVI TSRLSLVTQS VQQYINRIAL NLEPGLSIN QQEATDWEEF ANRYGYWAAN QQLRMFPEIY VDPTLRLTKT EFFFQLESAL NQGKLTDDVA QKAVLGYLNN F EEVSNLEI IAGYQDGIDI ENDKTYFVAR TRMQPYRYFW RSLDASQRNA NSQELYPTAW SEWKAISVPL ENVANGIVRP IM MDNRLYI SWFEVAEEKE TDSDGNIIVS GRYRTKIRLA HLGFDGVWSS GTTLREEVLA DQMEEMIAVV DRMEDEPRLA LVA FKEMSE SWDVVFSYIC DSMLIESSNL PTTTHPPKPG DGDKGLSDLD DYGANLVWFY LHETANGGKA EYKQLILYPV IINR DWPIE LDKTHQGDFG TVDDFTLNSN YTGDELSLYL QSSSTYKYDF SKSKNIIYGI WKEDANNNRC WLNYKLLTPE DYEPQ INAT LVMCDKGDVN IITGFSLPNG GVDAGGKIKV TLRVGKKLRD KFQIKQFSQT QYLQFPEASS ADVWYIGKQI RLNTLF AKE LIGKASRSLD LVLSWETQNS RLEEAILGGA AELIDLDGAN GIYFWELFFH MPFMVSWRFN VEQRYEDANR WVKYLFN PF ECEDEPALLL GKPPYWNSRP LVDEPFKGYS LTQPSDPDAI AASDPIHYRK AVFNFLTKNI IDQGDMEYRK LQPSARTL A RLSYSTASSL LGRRPDVQLT SFWQPLTLED ASYKTDSEIR AIEMQSQPLT FEPVVHDQTM SAVDNDIFMY PMNNELRGL WDRIENRIYN LRHNLTLDGK EINMDLYDSS ISPRGLMKQR YQRVVTARNA SKMNFKVPNY RFEPMLNRSK SGVETLIQFG STLLSLLER KDSLSFDAYQ MIQSGDLYRF SIDLQQQDID INKASLEALQ VSKQSAQDRY DHFKELYDEN ISSTEQKVIE L QSQAANSL LMAQGMRTAA AALDVIPNIY GLAVGGSHWG APLNAAAEII MIKYQADSSK SESLSVSESY RRRRQEWELQ YK QAEWEVN SVEQQINLQN MQIKAANKRL EQVEAQQQQA MALLDYFSER FTNESLYTWL ISQLSSLYLQ AYDAVLSLCL SAE ASLLYE LNLGEQSFVG GGGWNDLYQG LMAGETLKLA LMRMERVYVE QNSRRQEITK TISLKALLGE SWPAELNKLK QKTP INFNL EEQIFVEDYQ ELYQRRIKSV SVSLPMLVGP YEDVCAQLTQ TSSSYSTRAD LKTVENMLTK RTFADTPHLV RSIQP NQQI SLSTGVNDSG LFMLNFDDER FLPFEGSGVD SSWRLQFTNL KQNLDSLNDV ILHVKYTAAI GSSTFSQGVR KILANI NND E UniProtKB: Toxin subunit YenA2 |
-Macromolecule #3: Chitinase 2
| Macromolecule | Name: Chitinase 2 / type: protein_or_peptide / ID: 3 / Number of copies: 5 / Enantiomer: LEVO / EC number: chitinase |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Molecular weight | Theoretical: 69.740609 KDa |
| Sequence | String: MVNKYTYTSS KAMSDISDVI GEPLAAWDSQ VGGRVFNVIF DGKVYTNTYW VERWQVPGIG SSDGNPHNAW KFVRAATADE INKIGNPTT ADVKPTENIP SPILVEDKYT EETYSRPDVN FKEDGSQGNL SYTATRVCAP MYNHYVGDKT KPKLSAYITD W CQYDARLD ...String: MVNKYTYTSS KAMSDISDVI GEPLAAWDSQ VGGRVFNVIF DGKVYTNTYW VERWQVPGIG SSDGNPHNAW KFVRAATADE INKIGNPTT ADVKPTENIP SPILVEDKYT EETYSRPDVN FKEDGSQGNL SYTATRVCAP MYNHYVGDKT KPKLSAYITD W CQYDARLD GGGSKEEERG RGFDLATLMQ NPATYDRLIF SFLGICGDIG NKSKKVQEVW DGWNAQAPSL GLPQIGKGHI VP LDPYGDL GTARNVGLPP ESADTSIESG TFLPYYQQNR AAGLLGGLRE LQKKAHAMGH KLDLAFSIGG WSLSSYFSAL AEN PDERRV FVASVVDFFV RFPMFSCVDI DWEYPGGGGD EGNISSDKDG ENYVLLIKEL RSALDSRFGY SNRKEISIAC SGVK AKLKK SNIDQLVANG LDNIYLMSYD FFGTIWADYI GHHTNLYSPK DPGEQELFDL SAEAAIDYLH NELGIPMEKI HLGYA NYGR SAVGGDLTTR QYTKNGPALG TMENGAPEFF DIVKNYMDAE HSLSMGKNGF VLMTDTNADA DFLFSEAKGH FISLDT PRT VKQKGEYAAK NKLGGVFSWS GDQDCGLLAN AAREGLGYVA DSNQETIDMG PLYNPGKEIY LKSISEIKSK UniProtKB: Chitinase 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)