[English] 日本語
 Yorodumi
Yorodumi- EMDB-20054: Cryo-EM structure of YenTcA in the pore state determined in liposomes -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20054 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of YenTcA in the pore state determined in liposomes | |||||||||
 Map data Map data | em-volume_P1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) | |||||||||
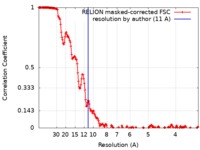
| Method | single particle reconstruction / cryo EM / Resolution: 11.0 Å | |||||||||
 Authors Authors | Landsberg MJ / Piper SJ | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin. Authors: Sarah J Piper / Lou Brillault / Rosalba Rothnagel / Tristan I Croll / Joseph K Box / Irene Chassagnon / Sebastian Scherer / Kenneth N Goldie / Sandra A Jones / Femke Schepers / Lauren ...Authors: Sarah J Piper / Lou Brillault / Rosalba Rothnagel / Tristan I Croll / Joseph K Box / Irene Chassagnon / Sebastian Scherer / Kenneth N Goldie / Sandra A Jones / Femke Schepers / Lauren Hartley-Tassell / Thomas Ve / Jason N Busby / Julie E Dalziel / J Shaun Lott / Ben Hankamer / Henning Stahlberg / Mark R H Hurst / Michael J Landsberg /      Abstract: ABC toxins are pore-forming virulence factors produced by pathogenic bacteria. YenTcA is the pore-forming and membrane binding A subunit of the ABC toxin YenTc, produced by the insect pathogen ...ABC toxins are pore-forming virulence factors produced by pathogenic bacteria. YenTcA is the pore-forming and membrane binding A subunit of the ABC toxin YenTc, produced by the insect pathogen Yersinia entomophaga. Here we present cryo-EM structures of YenTcA, purified from the native source. The soluble pre-pore structure, determined at an average resolution of 4.4 Å, reveals a pentameric assembly that in contrast to other characterised ABC toxins is formed by two TcA-like proteins (YenA1 and YenA2) and decorated by two endochitinases (Chi1 and Chi2). We also identify conformational changes that accompany membrane pore formation by visualising YenTcA inserted into liposomes. A clear outward rotation of the Chi1 subunits allows for access of the protruding translocation pore to the membrane. Our results highlight structural and functional diversity within the ABC toxin subfamily, explaining how different ABC toxins are capable of recognising diverse hosts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20054.map.gz emd_20054.map.gz | 27.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20054-v30.xml emd-20054-v30.xml emd-20054.xml emd-20054.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20054_fsc.xml emd_20054_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_20054.png emd_20054.png | 51 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20054 http://ftp.pdbj.org/pub/emdb/structures/EMD-20054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20054 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20054 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20054.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20054.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-volume_P1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.72 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Yersinia entomphaga toxin complex subunit A (YenTcA) in the pore ...
| Entire | Name: Yersinia entomphaga toxin complex subunit A (YenTcA) in the pore form in liposomes |
|---|---|
| Components |
|
-Supramolecule #1: Yersinia entomphaga toxin complex subunit A (YenTcA) in the pore ...
| Supramolecule | Name: Yersinia entomphaga toxin complex subunit A (YenTcA) in the pore form in liposomes type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) / Strain: MH96 Yersinia entomophaga (bacteria) / Strain: MH96 |
| Molecular weight | Theoretical: 2.08 MDa |
-Macromolecule #1: YenA1
| Macromolecule | Name: YenA1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Sequence | String: MDKYNNYSNV IKNKSSISPL LAAAAKIEPE ITVLSSASKS NRSQYSQSLA DTLLGLGYRS IFDIAKVSR QRFIKRHDES LLGNGAVIFD KAVSMANQVL QKYRKNRLEK SNSPLVPQTS S STDASSES QTNKLPEYNQ LFPEPWDNFC RPGAIEALDS PASYLLDLYK ...String: MDKYNNYSNV IKNKSSISPL LAAAAKIEPE ITVLSSASKS NRSQYSQSLA DTLLGLGYRS IFDIAKVSR QRFIKRHDES LLGNGAVIFD KAVSMANQVL QKYRKNRLEK SNSPLVPQTS S STDASSES QTNKLPEYNQ LFPEPWDNFC RPGAIEALDS PASYLLDLYK FIQSVELDGS NQ ARKLETR RADIPKLSLD NDALYKEVTA LSIVNDVLSG SAREYIDQSG QADKAVNQIL GDT HFPFTL PYSLPTQQIN KGLGASNIEL GTVIQRVDPQ FSWNTTQEKY NQVLLAYTQL SSEQ IALLS LPDVFTQNFL TQTELSAGYL SASTTEILAE KDLSRHGYIV KAADNIKGPT QLVEH SDAS YDVIELTCTN QAKETITVKL RGENIITYQR TKARMVPFDN SSPFSRQLKL TFVAED NPS LGNLDKGPYF ANMDIYAAEW VRENVSSETM VSRPFLTMTY RIAIAKAGAS LEELQPE AD AFFINNFGLS AEDSSQLVKL VAFGDQTGSK AEEIESLLSC GENLPIVSPN VIFANPIF G SYFNDEPFPA PYHFGGVYIN AHQRNAMTII RAEGGREIQS LSNFRLERLN RFIRLQRWL DLPSHQLDLL LTSVMQADAD NSQQEITEPV LKSLGLFRHL NLQYKITPEI FSSWLYQLTP FAVSGEIAF FDRIFNREQL FDQPFILDGG SFTYLDAKGS DAKSVKQLCA GLNISAVTFQ F IAPLVQSA LGLEAGTLVR SFEVVSSLYR LVSIPQTFGL STEDGLILMN ILTDEMGYLA KQ PAFDDKQ TQDKDFLSII LKMEALSAWL TKNNLTPASL ALLLGVTRLA VVPTNNMVTF FKG IANGLS ENVCLTTDDF QRQELEGADW WTLLSTNQVI DDMGLVLDIH PVWGKSDEEM LMEK IQSIG VSNDNNTLSI IVQILIQAKN AQENLLSQTI SAEYGVERSV VPLQLRWLGS NVYSV LNQV LNNTPTDISS IVPKLSELTY SLLIYTQLIN SLKLNKEFIF LRLTQPNWLG LTQPKL STQ LSLPEIYLIT CYQDWVVNAN KNEDSIHEYL EFANIKKTEA EKTLVDNSEK CAELLAE IL AWDAGEILKA ASLLGLNPPQ ATNVFEIDWI RRLQTLSEKT MISTEYLWQM GDLTENSE F SLKEGVGEAV MAALKAQGDS DNV |
-Macromolecule #2: YenA2
| Macromolecule | Name: YenA2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Sequence | String: MSNSIEAKLQ EDLRDALVDY YLGQIVPNSK DFTNLRSTIK NVDDLYDHLL LDTQVSAKVI TSRLSLVTQ SVQQYINRIA LNLEPGLSIN QQEATDWEEF ANRYGYWAAN QQLRMFPEIY V DPTLRLTK TEFFFQLESA LNQGKLTDDV AQKAVLGYLN NFEEVSNLEI ...String: MSNSIEAKLQ EDLRDALVDY YLGQIVPNSK DFTNLRSTIK NVDDLYDHLL LDTQVSAKVI TSRLSLVTQ SVQQYINRIA LNLEPGLSIN QQEATDWEEF ANRYGYWAAN QQLRMFPEIY V DPTLRLTK TEFFFQLESA LNQGKLTDDV AQKAVLGYLN NFEEVSNLEI IAGYQDGIDI EN DKTYFVA RTRMQPYRYF WRSLDASQRN ANSQELYPTA WSEWKAISVP LENVANGIVR PIM MDNRLY ISWFEVAEEK ETDSDGNIIV SGRYRTKIRL AHLGFDGVWS SGTTLREEVL ADQM EEMIA VVDRMEDEPR LALVAFKEMS ESWDVVFSYI CDSMLIESSN LPTTTHPPKP GDGDK GLSD LDDYGANLVW FYLHETANGG KAEYKQLILY PVIINRDWPI ELDKTHQGDF GTVDDF TLN SNYTGDELSL YLQSSSTYKY DFSKSKNIIY GIWKEDANNN RCWLNYKLLT PEDYEPQ IN ATLVMCDKGD VNIITGFSLP NGGVDAGGKI KVTLRVGKKL RDKFQIKQFS QTQYLQFP E ASSADVWYIG KQIRLNTLFA KELIGKASRS LDLVLSWETQ NSRLEEAILG GAAELIDLD GANGIYFWEL FFHMPFMVSW RFNVEQRYED ANRWVKYLFN PFECEDEPAL LLGKPPYWNS RPLVDEPFK GYSLTQPSDP DAIAASDPIH YRKAVFNFLT KNIIDQGDME YRKLQPSART L ARLSYSTA SSLLGRRPDV QLTSFWQPLT LEDASYKTDS EIRAIEMQSQ PLTFEPVVHD QT MSAVDND IFMYPMNNEL RGLWDRIENR IYNLRHNLTL DGKEINMDLY DSSISPRGLM KQR YQRVVT ARNASKMNFK VPNYRFEPML NRSKSGVETL IQFGSTLLSL LERKDSLSFD AYQM IQSGD LYRFSIDLQQ QDIDINKASL EALQVSKQSA QDRYDHFKEL YDENISSTEQ KVIEL QSQA ANSLLMAQGM RTAAAALDVI PNIYGLAVGG SHWGAPLNAA AEIIMIKYQA DSSKSE SLS VSESYRRRRQ EWELQYKQAE WEVNSVEQQI NLQNMQIKAA NKRLEQVEAQ QQQAMAL LD YFSERFTNES LYTWLISQLS SLYLQAYDAV LSLCLSAEAS LLYELNLGEQ SFVGGGGW N DLYQGLMAGE TLKLALMRME RVYVEQNSRR QEITKTISLK ALLGESWPAE LNKLKQKTP INFNLEEQIF VEDYQELYQR RIKSVSVSLP MLVGPYEDVC AQLTQTSSSY STRADLKTVE NMLTKRTFA DTPHLVRSIQ PNQQISLSTG VNDSGLFMLN FDDERFLPFE GSGVDSSWRL Q FTNLKQNL DSLNDVILHV KYTAAIGSST FSQGVRKILA NINNDE |
-Macromolecule #3: Chi1
| Macromolecule | Name: Chi1 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Sequence | String: MEKEEKSNLI YDKDPGYVWD NKNECEGAAE ETYQELNYEP SISADKLTWT PTRLAKTVFN TYEDDDDFN VLCYFTDWSQ YDPRIINKEI RDTGGRSADI LRLNTPDGRP FKRLIYSFGG L IGDKKYSA DGNASIAVRL GVATDPDDAI ANHKGKTIPV DPDGAVLASI ...String: MEKEEKSNLI YDKDPGYVWD NKNECEGAAE ETYQELNYEP SISADKLTWT PTRLAKTVFN TYEDDDDFN VLCYFTDWSQ YDPRIINKEI RDTGGRSADI LRLNTPDGRP FKRLIYSFGG L IGDKKYSA DGNASIAVRL GVATDPDDAI ANHKGKTIPV DPDGAVLASI NCGFTKWEAG DA NERYNQE KAKGLLGGFR LLHEADKELE FSLSIGGWSM SGLFSEIAKD EILRTNFVEG IKD FFQRFP MFSHLDIDWE YPGSIGAGNP NSPDDGANFA ILIQQITDAK ISNLKGISIA SSAD PAKID AANIPALMDA GVTGINLMTY DFFTLGDGKL SHHTNIYRDP SDVYSKYSID DAVTH LIDE KKVDPKAIFI GYAGYTRNAK NATITTSIPS EEALKGTYTD ANQTLGSFEY SVLEWT DII CHYMDFEKGE GRNGYKLVHD KVAKADYLYS EATKVFISLD TPRSVRDKGR YVKDKGL GG LFIWSGDQDN GILTNAAHEG LKRRIKNKVI DMTPFYLDSD EELPTYTEPA EPQCEACN I K |
-Macromolecule #4: Chi2
| Macromolecule | Name: Chi2 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Yersinia entomophaga (bacteria) Yersinia entomophaga (bacteria) |
| Sequence | String: MVNKYTYTSS KAMSDISDVI GEPLAAWDSQ VGGRVFNVIF DGKVYTNTYW VERWQVPGIG SSDGNPHNA WKFVRAATAD EINKIGNPTT ADVKPTENIP SPILVEDKYT EETYSRPDVN F KEDGSQGN LSYTATRVCA PMYNHYVGDK TKPKLSAYIT DWCQYDARLD ...String: MVNKYTYTSS KAMSDISDVI GEPLAAWDSQ VGGRVFNVIF DGKVYTNTYW VERWQVPGIG SSDGNPHNA WKFVRAATAD EINKIGNPTT ADVKPTENIP SPILVEDKYT EETYSRPDVN F KEDGSQGN LSYTATRVCA PMYNHYVGDK TKPKLSAYIT DWCQYDARLD GGGSKEEERG RG FDLATLM QNPATYDRLI FSFLGICGDI GNKSKKVQEV WDGWNAQAPS LGLPQIGKGH IVP LDPYGD LGTARNVGLP PESADTSIES GTFLPYYQQN RAAGLLGGLR ELQKKAHAMG HKLD LAFSI GGWSLSSYFS ALAENPDERR VFVASVVDFF VRFPMFSCVD IDWEYPGGGG DEGNI SSDK DGENYVLLIK ELRSALDSRF GYSNRKEISI ACSGVKAKLK KSNIDQLVAN GLDNIY LMS YDFFGTIWAD YIGHHTNLYS PKDPGEQELF DLSAEAAIDY LHNELGIPME KIHLGYA NY GRSAVGGDLT TRQYTKNGPA LGTMENGAPE FFDIVKNYMD AEHSLSMGKN GFVLMTDT N ADADFLFSEA KGHFISLDTP RTVKQKGEYA AKNKLGGVFS WSGDQDCGLL ANAAREGLG YVADSNQETI DMGPLYNPGK EIYLKSISEI KSK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)