[English] 日本語
 Yorodumi
Yorodumi- EMDB-1965: Structural and Functional Studies of LRP6 Ectodomain Reveal a Pla... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1965 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural and Functional Studies of LRP6 Ectodomain Reveal a Platform for Wnt Signaling | |||||||||
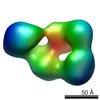
 Map data Map data | This is a surface rendering of the ectodomain of LRP6 bound to its chaperone Mesd | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | LDL-receptor-related protein 6 / Wnt signaling pathway / Wnt co-receptor | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 26.0 Å | |||||||||
 Authors Authors | Chen S / Bubeck D / MacDonald BT / Liang WX / Mao JH / Malinauskas T / Llorca O / Aricescu AR / Siebold C / He X / Jones EY | |||||||||
 Citation Citation |  Journal: Dev Cell / Year: 2011 Journal: Dev Cell / Year: 2011Title: Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Authors: Shuo Chen / Doryen Bubeck / Bryan T MacDonald / Wen-Xue Liang / Jian-Hua Mao / Tomas Malinauskas / Oscar Llorca / A Radu Aricescu / Christian Siebold / Xi He / E Yvonne Jones /  Abstract: LDL-receptor-related protein 6 (LRP6), alongside Frizzled receptors, transduces Wnt signaling across the plasma membrane. The LRP6 ectodomain comprises four tandem β-propeller-EGF-like domain (PE) ...LDL-receptor-related protein 6 (LRP6), alongside Frizzled receptors, transduces Wnt signaling across the plasma membrane. The LRP6 ectodomain comprises four tandem β-propeller-EGF-like domain (PE) pairs that harbor binding sites for Wnt morphogens and their antagonists including Dickkopf 1 (Dkk1). To understand how these multiple interactions are integrated, we combined crystallographic analysis of the third and fourth PE pairs with electron microscopy (EM) to determine the complete ectodomain structure. An extensive inter-pair interface, conserved for the first-to-second and third-to-fourth PE interactions, contributes to a compact platform-like architecture, which is disrupted by mutations implicated in developmental diseases. EM reconstruction of the LRP6 platform bound to chaperone Mesd exemplifies a binding mode spanning PE pairs. Cellular and binding assays identify overlapping Wnt3a- and Dkk1-binding surfaces on the third PE pair, consistent with steric competition, but also suggest a model in which the platform structure supports an interplay of ligands through multiple interaction sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1965.map.gz emd_1965.map.gz | 790.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1965-v30.xml emd-1965-v30.xml emd-1965.xml emd-1965.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1965_LRP6_MESD3.jpg 1965_LRP6_MESD3.jpg | 42.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1965 http://ftp.pdbj.org/pub/emdb/structures/EMD-1965 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1965 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1965 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1965.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1965.map.gz / Format: CCP4 / Size: 825.2 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a surface rendering of the ectodomain of LRP6 bound to its chaperone Mesd | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ectodomain of LRP6 residues 20 to 1361 and chaperone Mesd
| Entire | Name: Ectodomain of LRP6 residues 20 to 1361 and chaperone Mesd |
|---|---|
| Components |
|
-Supramolecule #1000: Ectodomain of LRP6 residues 20 to 1361 and chaperone Mesd
| Supramolecule | Name: Ectodomain of LRP6 residues 20 to 1361 and chaperone Mesd type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: one LRP6 binds to one Mesd / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 200 KDa / Method: Multi angle light scattering |
-Macromolecule #1: LDL-receptor-related protein 6
| Macromolecule | Name: LDL-receptor-related protein 6 / type: protein_or_peptide / ID: 1 / Name.synonym: LRP6 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Experimental: 100 KDa |
| Recombinant expression | Organism: HEK293S / Recombinant plasmid: pHLsec vector |
-Macromolecule #2: Mesoderm development protein
| Macromolecule | Name: Mesoderm development protein / type: protein_or_peptide / ID: 2 / Name.synonym: Mesd / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human Homo sapiens (human) / synonym: Human |
| Molecular weight | Experimental: 100 KDa |
| Recombinant expression | Organism: HEK293S / Recombinant plasmid: pHLsec vector |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.015 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 150 mM NaCl,10 mM Tris-HCl |
| Staining | Type: NEGATIVE Details: Grids were negatively stained with 0.75% uranyl formate using the two-drop method |
| Grid | Details: carbon-coated copper grids |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1230 |
|---|---|
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Sampling interval: 4.56 µm / Average electron dose: 10 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.9 mm / Nominal magnification: 72500 |
| Sample stage | Specimen holder: single-tilt / Specimen holder model: JEOL |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 26.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, XMIPP / Number images used: 7928 |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















