[English] 日本語
 Yorodumi
Yorodumi- EMDB-17692: Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tand... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | AMPAR / ion channels / neurotransmission / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationPresynaptic depolarization and calcium channel opening / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / channel regulator activity / LGI-ADAM interactions / Trafficking of AMPA receptors / regulation of AMPA receptor activity / membrane hyperpolarization ...Presynaptic depolarization and calcium channel opening / eye blink reflex / positive regulation of protein localization to basolateral plasma membrane / cerebellar mossy fiber / postsynaptic neurotransmitter receptor diffusion trapping / channel regulator activity / LGI-ADAM interactions / Trafficking of AMPA receptors / regulation of AMPA receptor activity / membrane hyperpolarization / nervous system process / protein targeting to membrane / voltage-gated calcium channel complex / neurotransmitter receptor localization to postsynaptic specialization membrane / neuromuscular junction development / AMPA glutamate receptor activity / response to lithium ion / transmission of nerve impulse / AMPA glutamate receptor complex / regulation of receptor recycling / membrane depolarization / regulation of postsynaptic membrane neurotransmitter receptor levels / positive regulation of synaptic transmission, glutamatergic / response to fungicide / voltage-gated calcium channel activity / somatodendritic compartment / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / ionotropic glutamate receptor binding / dendritic shaft / hippocampal mossy fiber to CA3 synapse / calcium channel regulator activity / regulation of membrane potential / response to calcium ion / postsynaptic density membrane / Schaffer collateral - CA1 synapse / amyloid-beta binding / dendritic spine / glutamatergic synapse / cell surface Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
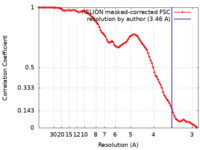
| Method | single particle reconstruction / cryo EM / Resolution: 3.46 Å | ||||||||||||
 Authors Authors | Zhang D / Krieger JM / Yamashita K / Greger IH | ||||||||||||
| Funding support |  United Kingdom, European Union, 3 items United Kingdom, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor. Authors: Danyang Zhang / Josip Ivica / James M Krieger / Hinze Ho / Keitaro Yamashita / Imogen Stockwell / Rozbeh Baradaran / Ondrej Cais / Ingo H Greger /   Abstract: AMPA glutamate receptors (AMPARs), the primary mediators of excitatory neurotransmission in the brain, are either GluA2 subunit-containing and thus Ca-impermeable, or GluA2-lacking and Ca-permeable. ...AMPA glutamate receptors (AMPARs), the primary mediators of excitatory neurotransmission in the brain, are either GluA2 subunit-containing and thus Ca-impermeable, or GluA2-lacking and Ca-permeable. Despite their prominent expression throughout interneurons and glia, their role in long-term potentiation and their involvement in a range of neuropathologies, structural information for GluA2-lacking receptors is currently absent. Here we determine and characterize cryo-electron microscopy structures of the GluA1 homotetramer, fully occupied with TARPγ3 auxiliary subunits (GluA1/γ3). The gating core of both resting and open-state GluA1/γ3 closely resembles GluA2-containing receptors. However, the sequence-diverse N-terminal domains (NTDs) give rise to a highly mobile assembly, enabling domain swapping and subunit re-alignments in the ligand-binding domain tier that are pronounced in desensitized states. These transitions underlie the unique kinetic properties of GluA1. A GluA2 mutant (F231A) increasing NTD dynamics phenocopies this behaviour, and exhibits reduced synaptic responses, reflecting the anchoring function of the AMPAR NTD at the synapse. Together, this work underscores how the subunit-diverse NTDs determine subunit arrangement, gating properties and ultimately synaptic signalling efficiency among AMPAR subtypes. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17692.map.gz emd_17692.map.gz | 32.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17692-v30.xml emd-17692-v30.xml emd-17692.xml emd-17692.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17692_fsc.xml emd_17692_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_17692.png emd_17692.png | 73.4 KB | ||
| Filedesc metadata |  emd-17692.cif.gz emd-17692.cif.gz | 6.6 KB | ||
| Others |  emd_17692_half_map_1.map.gz emd_17692_half_map_1.map.gz emd_17692_half_map_2.map.gz emd_17692_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17692 http://ftp.pdbj.org/pub/emdb/structures/EMD-17692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17692 | HTTPS FTP |
-Related structure data
| Related structure data |  8pivMC  8c1pC  8c1qC  8c1rC  8c1sC  8c2hC  8c2iC  8p3qC  8p3sC  8p3tC  8p3uC  8p3vC  8p3wC  8p3xC  8p3yC  8p3zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17692.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17692.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4455 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17692_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
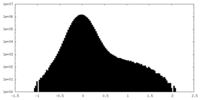
| Projections & Slices |
| ||||||||||||
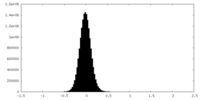
| Density Histograms |
-Half map: #1
| File | emd_17692_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
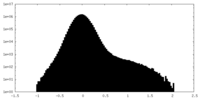
| Projections & Slices |
| ||||||||||||
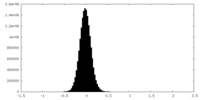
| Density Histograms |
- Sample components
Sample components
-Entire : GluA2 irR F231A mutant AMPA receptor (R/G unedited) in tandem wit...
| Entire | Name: GluA2 irR F231A mutant AMPA receptor (R/G unedited) in tandem with TARP gamma 2 in complex with 1mM quisqualate |
|---|---|
| Components |
|
-Supramolecule #1: GluA2 irR F231A mutant AMPA receptor (R/G unedited) in tandem wit...
| Supramolecule | Name: GluA2 irR F231A mutant AMPA receptor (R/G unedited) in tandem with TARP gamma 2 in complex with 1mM quisqualate type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: C-terminus of GluA2 flip-r-R with F231A is connected to gamma-2 N-terminus via a linker sequence |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glutamate receptor
| Macromolecule | Name: Glutamate receptor / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 98.984062 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQKIMHISVL LSPVLWGLIF GVSSNSIQIG GLFPRGADQE YSAFRVGMVQ FSTSEFRLTP HIDNLEVANS FAVTNAFCSQ FSRGVYAIF GFYDKKSVNT ITSFCGTLHV SFITPSFPTD GTHPFVIQMR PDLKGALLSL IEYYQWDKFA YLYDSDRGLS T LQAVLDSA ...String: MQKIMHISVL LSPVLWGLIF GVSSNSIQIG GLFPRGADQE YSAFRVGMVQ FSTSEFRLTP HIDNLEVANS FAVTNAFCSQ FSRGVYAIF GFYDKKSVNT ITSFCGTLHV SFITPSFPTD GTHPFVIQMR PDLKGALLSL IEYYQWDKFA YLYDSDRGLS T LQAVLDSA AEKKWQVTAI NVGNINNDKK DETYRSLFQD LELKKERRVI LDCERDKVND IVDQVITIGK HVKGYHYIIA NL GFTDGDL LKIQFGGANV SGFQIVDYDD SLVSKFIERW STLEEKEYPG AHTATIKYTS ALTYDAVQVM TEAFRNLRKQ RIE ISRRGN AGDCLANPAV PWGQGVEIER ALKQVQVEGL SGNIKFDQNG KRINYTINIM ELKTNGPRKI GYWSEVDKMV VTLT ELPSG NDTSGLENKT VVVTTILESP YVMMKKNHEM LEGNERYEGY CVDLAAEIAK HCGFKYKLTI VGDGKYGARD ADTKI WNGM VGELVYGKAD IAIAPLTITL VREEVIDFSK PFMSLGISIM IKKPQKSKPG VFSFLDPLAY EIWMCIVFAY IGVSVV LFL VSRFSPYEWH TEEFEDGRET QSSESTNEFG IFNSLWFSLG AFMRQGCDIS PRSLSGRIVG GVWWFFTLII ISSYTAN LA AFLTVERMVS PIESAEDLSK QTEIAYGTLD SGSTKEFFRR SKIAVFDKMW TYMRSAEPSV FVRTTAEGVA RVRKSKGK Y AYLLESTMNE YIEQRKPCDT MKVGGNLDSK GYGIATPKGS SLRTPVNLAV LKLSEQGVLD KLKNKWWYDK GECGAKDSG SKEKTSALSL SNVAGVFYIL VGGLGLAMLV ALIEFCYKSR AEAKRMKVAK NPQNINPSSS QNSQNFATYK EGYNVYGIES VKI UniProtKB: Glutamate receptor |
-Macromolecule #2: Voltage-dependent calcium channel gamma-2 subunit
| Macromolecule | Name: Voltage-dependent calcium channel gamma-2 subunit / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.882656 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGLFDRGVQM LLTTVGAFAA FSLMTIAVGT DYWLYSRGVC KTKSVSENET SKKNEEVMTH SGLWRTCCLE GNFKGLCKQI DHFPEDADY EADTAEYFLR AVRASSIFPI LSVILLFMGG LCIAASEFYK TRHNIILSAG IFFVSAGLSN IIGIIVYISA N AGDPSKSD ...String: MGLFDRGVQM LLTTVGAFAA FSLMTIAVGT DYWLYSRGVC KTKSVSENET SKKNEEVMTH SGLWRTCCLE GNFKGLCKQI DHFPEDADY EADTAEYFLR AVRASSIFPI LSVILLFMGG LCIAASEFYK TRHNIILSAG IFFVSAGLSN IIGIIVYISA N AGDPSKSD SKKNSYSYGW SFYFGALSFI IAEMVGVLAV HMFIDRHKQL TATARATDYL QASAITRIPS YRYRYQRRSR SS SRSTEPS HSRDASPVGV KGFNTLPSTE ISMYTLSRDP LKAATTPTAT YNSDRDNSFL QVHNCIQKDS KDSLHANTAN RRT TPV UniProtKB: Voltage-dependent calcium channel gamma-2 subunit |
-Macromolecule #3: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 3 / Number of copies: 7 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #4: (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate
| Macromolecule | Name: (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate / type: ligand / ID: 4 / Number of copies: 4 / Formula: OLC |
|---|---|
| Molecular weight | Theoretical: 356.54 Da |
| Chemical component information |  ChemComp-OLC: |
-Macromolecule #5: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 5 / Number of copies: 5 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 6 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)