[English] 日本語
 Yorodumi
Yorodumi- EMDB-16896: 39S mammalian mitochondrial large ribosomal subunit with mtRF1 an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 39S mammalian mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | ||||||||||||
 Map data Map data | 39S mammalian mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | mammalian mitochondrial ribosome / release factor mtRF1 / non-canonical stop codon / RIBOSOME | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial translation elongation / Mitochondrial translation termination / mitochondrial transcription / ribonuclease III activity / mitochondrial translational termination / mitochondrial translational elongation / translation release factor activity, codon nonspecific / Mitochondrial protein degradation / ribosomal subunit / translation release factor activity, codon specific ...Mitochondrial translation elongation / Mitochondrial translation termination / mitochondrial transcription / ribonuclease III activity / mitochondrial translational termination / mitochondrial translational elongation / translation release factor activity, codon nonspecific / Mitochondrial protein degradation / ribosomal subunit / translation release factor activity, codon specific / mitochondrial large ribosomal subunit / peptidyl-tRNA hydrolase / translation release factor activity / mitochondrial ribosome / peptidyl-tRNA hydrolase activity / mitochondrial translation / organelle membrane / RNA processing / rescue of stalled cytosolic ribosome / cellular response to leukemia inhibitory factor / cell junction / double-stranded RNA binding / large ribosomal subunit / mitochondrial inner membrane / negative regulation of translation / rRNA binding / nuclear body / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / protein domain specific binding / nucleotide binding / mRNA binding / positive regulation of DNA-templated transcription / mitochondrion / RNA binding / nucleoplasm / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
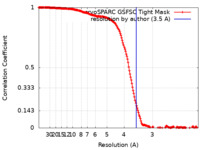
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Saurer M / Leibundgut M / Scaiola A / Schoenhut T / Ban N | ||||||||||||
| Funding support |  Switzerland, European Union, 3 items Switzerland, European Union, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Molecular basis of translation termination at noncanonical stop codons in human mitochondria. Authors: Martin Saurer / Marc Leibundgut / Hima Priyanka Nadimpalli / Alain Scaiola / Tanja Schönhut / Richard G Lee / Stefan J Siira / Oliver Rackham / René Dreos / Tea Lenarčič / Eva Kummer / ...Authors: Martin Saurer / Marc Leibundgut / Hima Priyanka Nadimpalli / Alain Scaiola / Tanja Schönhut / Richard G Lee / Stefan J Siira / Oliver Rackham / René Dreos / Tea Lenarčič / Eva Kummer / David Gatfield / Aleksandra Filipovska / Nenad Ban /    Abstract: The genetic code that specifies the identity of amino acids incorporated into proteins during protein synthesis is almost universally conserved. Mitochondrial genomes feature deviations from the ...The genetic code that specifies the identity of amino acids incorporated into proteins during protein synthesis is almost universally conserved. Mitochondrial genomes feature deviations from the standard genetic code, including the reassignment of two arginine codons to stop codons. The protein required for translation termination at these noncanonical stop codons to release the newly synthesized polypeptides is not currently known. In this study, we used gene editing and ribosomal profiling in combination with cryo-electron microscopy to establish that mitochondrial release factor 1 (mtRF1) detects noncanonical stop codons in human mitochondria by a previously unknown mechanism of codon recognition. We discovered that binding of mtRF1 to the decoding center of the ribosome stabilizes a highly unusual conformation in the messenger RNA in which the ribosomal RNA participates in specific recognition of the noncanonical stop codons. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16896.map.gz emd_16896.map.gz | 450.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16896-v30.xml emd-16896-v30.xml emd-16896.xml emd-16896.xml | 86.9 KB 86.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16896_fsc.xml emd_16896_fsc.xml | 16.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16896.png emd_16896.png | 195.1 KB | ||
| Filedesc metadata |  emd-16896.cif.gz emd-16896.cif.gz | 18.9 KB | ||
| Others |  emd_16896_half_map_1.map.gz emd_16896_half_map_1.map.gz emd_16896_half_map_2.map.gz emd_16896_half_map_2.map.gz | 443 MB 443 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16896 http://ftp.pdbj.org/pub/emdb/structures/EMD-16896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16896 | HTTPS FTP |
-Related structure data
| Related structure data |  8oiqMC  8oinC  8oipC  8oirC  8oisC  8oitC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16896.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16896.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 39S mammalian mitochondrial large ribosomal subunit with mtRF1 and P-site tRNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||
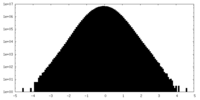
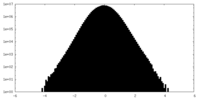
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_16896_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
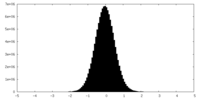
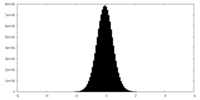
| Density Histograms |
-Half map: half map B
| File | emd_16896_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : 39S mammalian mitochondrial ribosome with mtRF1 and P-site tRNAMet
+Supramolecule #1: 39S mammalian mitochondrial ribosome with mtRF1 and P-site tRNAMet
+Macromolecule #1: Mitochondrial ribosomal protein L12
+Macromolecule #5: Large ribosomal subunit protein uL22m
+Macromolecule #6: 39S ribosomal protein L23, mitochondrial
+Macromolecule #7: uL24m
+Macromolecule #8: 39S ribosomal protein L27, mitochondrial
+Macromolecule #9: Mitochondrial ribosomal protein L28
+Macromolecule #10: Mitochondrial ribosomal protein L47
+Macromolecule #11: Large ribosomal subunit protein uL30m
+Macromolecule #12: Mitochondrial ribosomal protein L32
+Macromolecule #13: Large ribosomal subunit protein bL33m
+Macromolecule #14: Large ribosomal subunit protein bL34m
+Macromolecule #15: 39S ribosomal protein L35, mitochondrial
+Macromolecule #16: Mitochondrial ribosomal protein L2
+Macromolecule #17: ICT1
+Macromolecule #18: Mitochondrial ribosomal protein L4
+Macromolecule #19: Mitochondrial ribosomal protein L9
+Macromolecule #20: Mitochondrial ribosomal protein L10
+Macromolecule #21: Mitochondrial ribosomal protein L11
+Macromolecule #22: 39S ribosomal protein L13, mitochondrial
+Macromolecule #23: Mitochondrial ribosomal protein L14
+Macromolecule #24: 39S ribosomal protein L15, mitochondrial
+Macromolecule #25: uL16m
+Macromolecule #26: bL17m
+Macromolecule #27: Mitochondrial ribosomal protein L18
+Macromolecule #28: 39S ribosomal protein L19, mitochondrial
+Macromolecule #29: Mitochondrial ribosomal protein L20
+Macromolecule #30: Mitochondrial ribosomal protein L21
+Macromolecule #31: 39S ribosomal protein L52, mitochondrial
+Macromolecule #32: Mitochondrial ribosomal protein L53
+Macromolecule #33: mL54
+Macromolecule #34: Mitochondrial ribosomal protein L55
+Macromolecule #35: Mitochondrial ribosomal protein L57
+Macromolecule #36: mL62 (ICT1)
+Macromolecule #37: mL64
+Macromolecule #38: Mitochondrial ribosomal protein S18A
+Macromolecule #39: 39S ribosomal protein S30, mitochondrial
+Macromolecule #40: 39S ribosomal protein L1, mitochondrial
+Macromolecule #41: Ribosomal protein
+Macromolecule #42: Mitochondrial ribosomal protein L37
+Macromolecule #43: Mitochondrial ribosomal protein L38
+Macromolecule #44: Mitochondrial ribosomal protein L39
+Macromolecule #45: Mitochondrial ribosomal protein L40
+Macromolecule #46: 39S ribosomal protein L41, mitochondrial
+Macromolecule #47: mL42
+Macromolecule #48: Large ribosomal subunit protein mL43
+Macromolecule #49: mL44
+Macromolecule #50: Mitochondrial ribosomal protein L45
+Macromolecule #51: Mitochondrial ribosomal protein L46
+Macromolecule #52: 39S ribosomal protein L48, mitochondrial
+Macromolecule #53: 39S ribosomal protein L49, mitochondrial
+Macromolecule #54: Mitochondrial ribosomal protein L50
+Macromolecule #55: Large ribosomal subunit protein mL51
+Macromolecule #57: Peptide chain release factor 1, mitochondrial
+Macromolecule #2: E-site tRNA
+Macromolecule #3: 16S rRNA
+Macromolecule #4: CP Phe-tRNA
+Macromolecule #56: P-site Met-tRNA(fMet)
+Macromolecule #58: MAGNESIUM ION
+Macromolecule #59: POTASSIUM ION
+Macromolecule #60: PHENYLALANINE
+Macromolecule #61: ZINC ION
+Macromolecule #62: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #63: N-FORMYLMETHIONINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 20mM HEPES-KOH pH=7.5, 100mM KCl, 20mM MgCl2, ~1% sucrose (from mtRF1 storage buffer) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: CARBON / Support film - #0 - topology: HOLEY / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | In vitro reconstituted complex (55S). Focused refinement on large subunit. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6973 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||||||
| Output model |  PDB-8oiq: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)