[English] 日本語
 Yorodumi
Yorodumi- EMDB-15902: S. cerevisiae replisome + Ctf4 + pol alpha. Unbinned, locally fil... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae replisome + Ctf4 + pol alpha. Unbinned, locally filtered. | |||||||||
 Map data Map data | Yeast replisome pol alpha Ctf4. Unbinned, locally filtered. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Replication / helicase / polymerase / pol alpha / priming | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Jones ML / Yeeles JTP | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: How Pol α-primase is targeted to replisomes to prime eukaryotic DNA replication. Authors: Morgan L Jones / Valentina Aria / Yasemin Baris / Joseph T P Yeeles /  Abstract: During eukaryotic DNA replication, Pol α-primase generates primers at replication origins to start leading-strand synthesis and every few hundred nucleotides during discontinuous lagging-strand ...During eukaryotic DNA replication, Pol α-primase generates primers at replication origins to start leading-strand synthesis and every few hundred nucleotides during discontinuous lagging-strand replication. How Pol α-primase is targeted to replication forks to prime DNA synthesis is not fully understood. Here, by determining cryoelectron microscopy (cryo-EM) structures of budding yeast and human replisomes containing Pol α-primase, we reveal a conserved mechanism for the coordination of priming by the replisome. Pol α-primase binds directly to the leading edge of the CMG (CDC45-MCM-GINS) replicative helicase via a complex interaction network. The non-catalytic PRIM2/Pri2 subunit forms two interfaces with CMG that are critical for in vitro DNA replication and yeast cell growth. These interactions position the primase catalytic subunit PRIM1/Pri1 directly above the exit channel for lagging-strand template single-stranded DNA (ssDNA), revealing why priming occurs efficiently only on the lagging-strand template and elucidating a mechanism for Pol α-primase to overcome competition from RPA to initiate primer synthesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15902.map.gz emd_15902.map.gz | 28.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15902-v30.xml emd-15902-v30.xml emd-15902.xml emd-15902.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15902_fsc.xml emd_15902_fsc.xml | 19.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_15902.png emd_15902.png | 101.3 KB | ||
| Others |  emd_15902_half_map_1.map.gz emd_15902_half_map_1.map.gz emd_15902_half_map_2.map.gz emd_15902_half_map_2.map.gz | 690.9 MB 691 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15902 http://ftp.pdbj.org/pub/emdb/structures/EMD-15902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15902 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15902.map.gz / Format: CCP4 / Size: 744.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15902.map.gz / Format: CCP4 / Size: 744.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast replisome pol alpha Ctf4. Unbinned, locally filtered. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
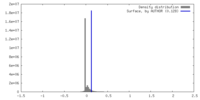
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Yeast replisome pol alpha Ctf4. Unbinned, locally filtered....
| File | emd_15902_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast replisome pol alpha Ctf4. Unbinned, locally filtered. Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
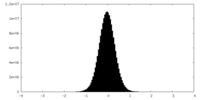
| Density Histograms |
-Half map: Yeast replisome pol alpha Ctf4. Unbinned, locally filtered....
| File | emd_15902_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast replisome pol alpha Ctf4. Unbinned, locally filtered. Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
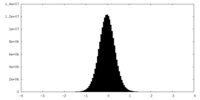
| Density Histograms |
- Sample components
Sample components
-Entire : Replisome - pol alpha complex
| Entire | Name: Replisome - pol alpha complex |
|---|---|
| Components |
|
-Supramolecule #1: Replisome - pol alpha complex
| Supramolecule | Name: Replisome - pol alpha complex / type: complex / ID: 1 / Parent: 0 Details: S. cerevisiae pol alpha bound to the core replisome engaged with a fork DNA substrate containing a 60 nucleotide lagging strand. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.184 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)