+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail of phage SU10 genome release intermediate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage / tail / base plate / nozzle / tail fibers / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationPhage tail protein C-terminal domain / : / SU10 adaptor protein / : / Baseplate structural protein Gp10, C-terminal domain / : / Phage SU10 portal protein / Bacterial Ig-like domain (group 2) / Invasin/intimin cell-adhesion fragments / Bacterial Ig-like domain 2 / Bacterial Ig-like domain, group 2 Similarity search - Domain/homology | |||||||||
| Biological species |  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) | |||||||||
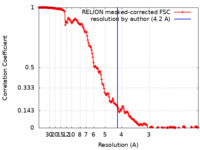
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Siborova M / Fuzik T / Prochazkova M / Novacek J / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Tail proteins of phage SU10 reorganize into the nozzle for genome delivery. Authors: Marta Šiborová / Tibor Füzik / Michaela Procházková / Jiří Nováček / Martin Benešík / Anders S Nilsson / Pavel Plevka /   Abstract: Escherichia coli phage SU10 belongs to the genus Kuravirus from the class Caudoviricetes of phages with short non-contractile tails. In contrast to other short-tailed phages, the tails of Kuraviruses ...Escherichia coli phage SU10 belongs to the genus Kuravirus from the class Caudoviricetes of phages with short non-contractile tails. In contrast to other short-tailed phages, the tails of Kuraviruses elongate upon cell attachment. Here we show that the virion of SU10 has a prolate head, containing genome and ejection proteins, and a tail, which is formed of portal, adaptor, nozzle, and tail needle proteins and decorated with long and short fibers. The binding of the long tail fibers to the receptors in the outer bacterial membrane induces the straightening of nozzle proteins and rotation of short tail fibers. After the re-arrangement, the nozzle proteins and short tail fibers alternate to form a nozzle that extends the tail by 28 nm. Subsequently, the tail needle detaches from the nozzle proteins and five types of ejection proteins are released from the SU10 head. The nozzle with the putative extension formed by the ejection proteins enables the delivery of the SU10 genome into the bacterial cytoplasm. It is likely that this mechanism of genome delivery, involving the formation of the tail nozzle, is employed by all Kuraviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14495.map.gz emd_14495.map.gz | 31 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14495-v30.xml emd-14495-v30.xml emd-14495.xml emd-14495.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14495_fsc.xml emd_14495_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14495.png emd_14495.png | 81.1 KB | ||
| Filedesc metadata |  emd-14495.cif.gz emd-14495.cif.gz | 8 KB | ||
| Others |  emd_14495_half_map_1.map.gz emd_14495_half_map_1.map.gz emd_14495_half_map_2.map.gz emd_14495_half_map_2.map.gz | 408.5 MB 408 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14495 http://ftp.pdbj.org/pub/emdb/structures/EMD-14495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14495 | HTTPS FTP |
-Related structure data
| Related structure data |  7z4fMC  7z44C  7z45C  7z46C  7z47C  7z48C  7z49C  7z4aC  7z4bC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14495.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14495.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14495_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
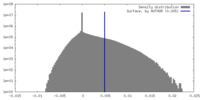
| Density Histograms |
-Half map: #1
| File | emd_14495_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
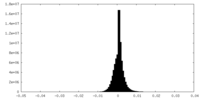
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia phage vB_EcoP_SU10
| Entire | Name:  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia phage vB_EcoP_SU10
| Supramolecule | Name: Escherichia phage vB_EcoP_SU10 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1519788 / Sci species name: Escherichia phage vB_EcoP_SU10 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
| Molecular weight | Theoretical: 3.036 MDa |
| Virus shell | Shell ID: 1 / Name: Tail complex |
-Macromolecule #1: Putative structural protein
| Macromolecule | Name: Putative structural protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) |
| Molecular weight | Theoretical: 29.258613 KDa |
| Sequence | String: MAIETNAVVI TDLNPLYPRD RDYIYEGAAQ IRLIKQTLQN TFPNVTEPVD IDSDTFKIMS EKLKFTGDAM DVGGLMIKNV TPGTGDKDV VTKGQMEAFM KNWMENKLYR IGSYYITEED INPGDSISLG FGSWAKVTGV IMGTGVVNPD GSVPNAQRVE F QAGGTGGR ...String: MAIETNAVVI TDLNPLYPRD RDYIYEGAAQ IRLIKQTLQN TFPNVTEPVD IDSDTFKIMS EKLKFTGDAM DVGGLMIKNV TPGTGDKDV VTKGQMEAFM KNWMENKLYR IGSYYITEED INPGDSISLG FGSWAKVTGV IMGTGVVNPD GSVPNAQRVE F QAGGTGGR VFNTIRTENV PLMTVNGSSF SLSSNTHSHN MVFGRGDASG HNSSPNWYSP GGGYSQRTDN DTHTHTISGS VS LGRDDIS RQPINTLPPF RAAHIWRRIS UniProtKB: Putative structural protein |
-Macromolecule #2: Putative tail fiber
| Macromolecule | Name: Putative tail fiber / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) |
| Molecular weight | Theoretical: 83.558227 KDa |
| Sequence | String: MIVYNNQAPD AVNNVGQFGA TEGSIGAYKQ AAEYAADSKY WALLAESKFG TIDDLIAEVE RLYQQGVLMK QDIEDLKQDF KDQDARLMS LIAQTNAAVS DANNAVALIN QKLIEVQNQL DVLLGMSVDV TTLPPGTPAT GSFNPNTGVI SLGIPEGEPG K DGSVKDLD ...String: MIVYNNQAPD AVNNVGQFGA TEGSIGAYKQ AAEYAADSKY WALLAESKFG TIDDLIAEVE RLYQQGVLMK QDIEDLKQDF KDQDARLMS LIAQTNAAVS DANNAVALIN QKLIEVQNQL DVLLGMSVDV TTLPPGTPAT GSFNPNTGVI SLGIPEGEPG K DGSVKDLD TAPTGVPELG DLGFYVDKDD NTVHKTTLEN IANLTPSVRS VSVNGGPALD GEVALTINKE TVGLGNVLNV AQ YSRQEIN DKFDKTTKTY QSKAEAYADA QYRQVGEKVL VWEATKYEFY TVAANKTLTP VKTEGRILTV NSRSPDSSGN IDI TIPTGN PSLYLGEMVM FPYDPSKNIS YPGVLPADGR LVSKESASDL GPSLVSGQLP VVSETEWQSG AKQYFSWGKL ADGI TDADS TNFINIRLPD WTGGEAIRAP DSDKDSQYNG SVQAQKPYVV TVNNQAPDEI TGNVNISRSI LGAASSGANS DITSL SGLT TPLSISQGGT GAKDAASARS NLGLGSVSTL DNVPIASGGT GAGDAAGARF NLGLGNSATM NTGTNSDNVL KVGDFG IGR PDGALVFDTT SQDQLLAGLD TYGLCVFRNN QQIAAPWDIW NYSSNLFFRA GDTYSMISIP FESAGKIKVF GGASGSG WK TSRTVYDTVN TTVDVNGFIK AASPIVKVFH DGSFETNEQS DGVSVKKIST GVYLISGCLG LNSDAGWGGV DGGFEIPI D RNKQPRVWLD YEVKEDGSLL IKTYHRTHST SPAFARNELE GFSDGDPVDI PKDAFISVRV EMPSK UniProtKB: Putative tail fiber |
-Macromolecule #3: Adaptor protein
| Macromolecule | Name: Adaptor protein / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) |
| Molecular weight | Theoretical: 28.836395 KDa |
| Sequence | String: MAMPDVQYPI NTYGWLKKAV ALWADRDDDE FVNQIPNFIN FAEKEIYRNL RIPPLEKEVY LDIKDGVAYI PPDYLEAQWM MRAKDGTIF QVTSPEEISY RRQHGTINPS HWNNQPVNFA RFGSRFIFYP SIEADTPYYP DDGSPLIPAE NSVILSYYAD P PEFHEDTD ...String: MAMPDVQYPI NTYGWLKKAV ALWADRDDDE FVNQIPNFIN FAEKEIYRNL RIPPLEKEVY LDIKDGVAYI PPDYLEAQWM MRAKDGTIF QVTSPEEISY RRQHGTINPS HWNNQPVNFA RFGSRFIFYP SIEADTPYYP DDGSPLIPAE NSVILSYYAD P PEFHEDTD TSTILTIAPE LLLYFTLRHA CLFVQDDNGV QKWSALGKAI LDEMVEQNKK QEYSGSPIAI PNNMTRLQSS LP DIYGIRT SRV UniProtKB: Uncharacterized protein |
-Macromolecule #4: Surface protein
| Macromolecule | Name: Surface protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) |
| Molecular weight | Theoretical: 110.352617 KDa |
| Sequence | String: MALYPIKSLG AVGVIADQAP TDLAPNAFTN AINARFVEQR VFKTGGNAPL SYVDEDKDLT PLSFVSMPFD YYSAGNSFLV VGTNKKLYK LTDESLTDIS RKVATVTKKA SASIKIYPVV SQIVPKESTI SMNFNQTKNL EVSLLPADAN NTNLIWEVSN S SYGSITVD ...String: MALYPIKSLG AVGVIADQAP TDLAPNAFTN AINARFVEQR VFKTGGNAPL SYVDEDKDLT PLSFVSMPFD YYSAGNSFLV VGTNKKLYK LTDESLTDIS RKVATVTKKA SASIKIYPVV SQIVPKESTI SMNFNQTKNL EVSLLPADAN NTNLIWEVSN S SYGSITVD PSDSKLATLT SFEKEGNLVV TISTANESVV AQIAVNIIDG DSGIFLSQDT VTIRKGGTTT LTAVTGKTPV TW SSNNASI VSVTPNANSL TAVITANGEG NVTITADNGT KTASCEIVSI PQIDSISLSQ SDVTVSRGSQ YILTATLSPA NAP NQNITW TSSNPNIATV SGTSTQGTIN ALLAGFTEIT ATTEEGNRVA VCTVRVDLAG RTMRTSAMAF AAPVSESVET QEEE VVTPP ESEETVYFAE PTSGIDTSGM YEGNNFYDYS NVNDIEGFAR ASLLATPLSS VTLDIVSASL DVGEEIVITA TASPE GEYS YQWSVDKTGY VSTTSVTGKS IKLVALRKGE INVTCTVSQM TQKDYDAFDD YPWYHAVISN CAVATTHYET PQVKEF ESE YFVDLPGWGE QTVVDNDGNP SVKKFNWKCE RVRSFNNRLF ALNMREANAS GVTTNYPLRL RWSNFANENK APTLWDD FA YDRVVSSDLA SNIVGQTQAL ENGYAGYIDL ADSNGSLIDI LPLKDYLFVY TEFETYIGSP TNNTYQPLMF KKLFNDSG I LAPECVVEVE GSHFVVTQND VILHNGATKK SIASNRVKNM LINEVCLVNP LATRVHLHQD KKEVWVLYVG PGEPKESFA CTKAAVWNYE FDTWSFRTIP YAQCIGLVDP PVLERGPIWS DFQEITWDDP SIKELVWRKD ATNFRQRVTI VGSFLKGFYQ VDVGALDYF YDRLNDVVIE KPLEMRLERT GIDFDNVTNE WNQKHINRFR PQTTGSGTYI FEAGGSQFSN EYGHPHTSKT Y TIGVDRHV SVRLNHPYLF YNVIDNDVNS NAAINGLTIE FAVGGRR UniProtKB: Surface protein |
-Macromolecule #5: Portal protein
| Macromolecule | Name: Portal protein / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage vB_EcoP_SU10 (virus) Escherichia phage vB_EcoP_SU10 (virus) |
| Molecular weight | Theoretical: 85.055344 KDa |
| Sequence | String: MAKQKYSEEV LDELRVDLQR RFNYAQGYVD MAVKGYAREA WEYFYGNLPA PVTAGSSSWV DRTVWESVNG TLQDIINVFC SGDEAVTFV ADNQQDSDAA DVATKLVNQI LLRDNPGYNI ISSAAQECLV TRNSFIKYYW DEQTSTQTEE AEGVPPEALA A YVQGLEAG ...String: MAKQKYSEEV LDELRVDLQR RFNYAQGYVD MAVKGYAREA WEYFYGNLPA PVTAGSSSWV DRTVWESVNG TLQDIINVFC SGDEAVTFV ADNQQDSDAA DVATKLVNQI LLRDNPGYNI ISSAAQECLV TRNSFIKYYW DEQTSTQTEE AEGVPPEALA A YVQGLEAG GLKNLEVFTE ENEDGTVDVK VTYEQTVKRV KVEYVPSEQI FVDEHATSFA DAQYFCHRVR RSKEDLVAMG FP KDEIEAF NDWTDTMDTT QSTVAWSRTD WRQDIDADIG TDTEDIASMV WVYEHYIRTG VLDKNKESKL YQVIQAGEHI LHT EEVTHI PFVTFCPYPI PGSFYGQSVY DITKDIQDLR TALVRGYIDN VNNANYGRYK ALVGAYDRRS LLDNRPGGVV EMER QDAID LFPYHNLPQG IDGLLGMSEE LKETRTGVTK LGMGINPDVF KNDNAYATVG LMMNAAQNRL RMVCRNIAHN GMVEL MRGI YNLIRENGEV PIEVQTPRGM IQVNPKQLPA RHNLQVVVAI SPNEKAERAQ KLISLKQLIA ADAQLAPLFG LEQDRY MTA QIFELMGIKD THKYLLPLEQ YQPPEPSPME ILQLEMTKAQ VENVQASSQK MIADAFDQRE RTTFEQQKAA DELSLRQ EE LQFKQENAAD AMTLENRKED NNATLEQAKH KLALMQQQVR QYESVLKELQ MVMSHQVDQE KIVQQARVQD KTLELQKK E ANVTKKEQQA SLKDSRIPGK RLGSKK UniProtKB: Portal protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | PFU 10^11 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 1.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
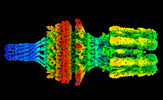
| Output model |  PDB-7z4f: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)