[English] 日本語
 Yorodumi
Yorodumi- EMDB-1428: A lever-arm rotation drives motility of the minus-end-directed ki... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1428 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. | |||||||||
 Map data Map data | Ncd in its ATP bound, AMPPNP, state on 15 filament microtubules | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 18.0 Å | |||||||||
 Authors Authors | Endres NF / Yoshioka C / Milligan RA / Vale RD | |||||||||
 Citation Citation |  Journal: Nature / Year: 2006 Journal: Nature / Year: 2006Title: A lever-arm rotation drives motility of the minus-end-directed kinesin Ncd. Authors: Nicholas F Endres / Craig Yoshioka / Ronald A Milligan / Ronald D Vale /  Abstract: Kinesins are microtubule-based motor proteins that power intracellular transport. Most kinesin motors, exemplified by Kinesin-1, move towards the microtubule plus end, and the structural changes that ...Kinesins are microtubule-based motor proteins that power intracellular transport. Most kinesin motors, exemplified by Kinesin-1, move towards the microtubule plus end, and the structural changes that govern this directional preference have been described. By contrast, the nature and timing of the structural changes underlying the minus-end-directed motility of Kinesin-14 motors (such as Drosophila Ncd) are less well understood. Using cryo-electron microscopy, here we demonstrate that a coiled-coil mechanical element of microtubule-bound Ncd rotates approximately 70 degrees towards the minus end upon ATP binding. Extending or shortening this coiled coil increases or decreases velocity, respectively, without affecting ATPase activity. An unusual Ncd mutant that lacks directional preference shows unstable nucleotide-dependent conformations of its coiled coil, underscoring the role of this mechanical element in motility. These results show that the force-producing conformational change in Ncd occurs on ATP binding, as in other kinesins, but involves the swing of a lever-arm mechanical element similar to that described for myosins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1428.map.gz emd_1428.map.gz | 513.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1428-v30.xml emd-1428-v30.xml emd-1428.xml emd-1428.xml | 8.9 KB 8.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1428.gif 1428.gif | 51.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1428 http://ftp.pdbj.org/pub/emdb/structures/EMD-1428 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1428 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1428 | HTTPS FTP |
-Validation report
| Summary document |  emd_1428_validation.pdf.gz emd_1428_validation.pdf.gz | 298.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1428_full_validation.pdf.gz emd_1428_full_validation.pdf.gz | 297.2 KB | Display | |
| Data in XML |  emd_1428_validation.xml.gz emd_1428_validation.xml.gz | 4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1428 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1428 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1428 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1428 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1428.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1428.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ncd in its ATP bound, AMPPNP, state on 15 filament microtubules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
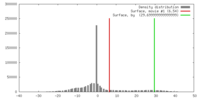
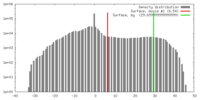
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : non-claret disjunction atp, amppnp, state
| Entire | Name: non-claret disjunction atp, amppnp, state |
|---|---|
| Components |
|
-Supramolecule #1000: non-claret disjunction atp, amppnp, state
| Supramolecule | Name: non-claret disjunction atp, amppnp, state / type: sample / ID: 1000 Oligomeric state: Motors bound to 15 protofilament helical microtubules Number unique components: 2 |
|---|
-Macromolecule #1: tubulin
| Macromolecule | Name: tubulin / type: protein_or_peptide / ID: 1 / Oligomeric state: helical 15 protofilaments / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #2: ncd
| Macromolecule | Name: ncd / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 25 mM MOPS (pH 7.25), 100 mM NaCl, 2 mM MgCl2, 1 mM EGTA and 1 mM dithiothreitol, 5mg/ml tubulin, 5mM AMPPNP |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 4 K / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: 3 sec blot |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 120 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal magnification: 39000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: OTHER / Software - Name: phoelix |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)