[English] 日本語
 Yorodumi
Yorodumi- EMDB-2043: Subtomogram averaging reconstruction of the Ebola virus nucleocapsid -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2043 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
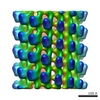
| Title | Subtomogram averaging reconstruction of the Ebola virus nucleocapsid | |||||||||
 Map data Map data | Reconstruction of the Ebola virus nucleocapsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ebola virus / nucleocapsid / subtomogram averaging | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 36.0 Å | |||||||||
 Authors Authors | Bharat TAM / Noda T / Riches JD / Kraehling V / Kolesnikova L / Becker S / Kawaoka Y / Briggs JAG | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2012 Journal: Proc Natl Acad Sci U S A / Year: 2012Title: Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Authors: Tanmay A M Bharat / Takeshi Noda / James D Riches / Verena Kraehling / Larissa Kolesnikova / Stephan Becker / Yoshihiro Kawaoka / John A G Briggs /  Abstract: Ebola virus is a highly pathogenic filovirus causing severe hemorrhagic fever with high mortality rates. It assembles heterogenous, filamentous, enveloped virus particles containing a negative-sense, ...Ebola virus is a highly pathogenic filovirus causing severe hemorrhagic fever with high mortality rates. It assembles heterogenous, filamentous, enveloped virus particles containing a negative-sense, single-stranded RNA genome packaged within a helical nucleocapsid (NC). We have used cryo-electron microscopy and tomography to visualize Ebola virus particles, as well as Ebola virus-like particles, in three dimensions in a near-native state. The NC within the virion forms a left-handed helix with an inner nucleoprotein layer decorated with protruding arms composed of VP24 and VP35. A comparison with the closely related Marburg virus shows that the N-terminal region of nucleoprotein defines the inner diameter of the Ebola virus NC, whereas the RNA genome defines its length. Binding of the nucleoprotein to RNA can assemble a loosely coiled NC-like structure; the loose coil can be condensed by binding of the viral matrix protein VP40 to the C terminus of the nucleoprotein, and rigidified by binding of VP24 and VP35 to alternate copies of the nucleoprotein. Four proteins (NP, VP24, VP35, and VP40) are necessary and sufficient to mediate assembly of an NC with structure, symmetry, variability, and flexibility indistinguishable from that in Ebola virus particles released from infected cells. Together these data provide a structural and architectural description of Ebola virus and define the roles of viral proteins in its structure and assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2043.map.gz emd_2043.map.gz | 612.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2043-v30.xml emd-2043-v30.xml emd-2043.xml emd-2043.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  2043_emd_2043.jpg 2043_emd_2043.jpg emd_2043.jpg emd_2043.jpg | 73.2 KB 98.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2043 http://ftp.pdbj.org/pub/emdb/structures/EMD-2043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2043 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2043.map.gz / Format: CCP4 / Size: 646.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2043.map.gz / Format: CCP4 / Size: 646.5 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the Ebola virus nucleocapsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 9.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ebola virus nucleocapsid
| Entire | Name: Ebola virus nucleocapsid |
|---|---|
| Components |
|
-Supramolecule #1000: Ebola virus nucleocapsid
| Supramolecule | Name: Ebola virus nucleocapsid / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Zaire ebolavirus
| Supramolecule | Name: Zaire ebolavirus / type: virus / ID: 1 / NCBI-ID: 186538 / Sci species name: Zaire ebolavirus / Database: NCBI / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: PBS buffer with 4% paraformaldehyde |
|---|---|
| Grid | Details: 300 mesh holey carbon C-Flat grid with 2micron holes. |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at high magnification. |
| Specialist optics | Energy filter - Name: Gatan (GIF 2002) |
| Date | Jun 30, 2011 |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN / Average electron dose: 80 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: -6.0 µm / Nominal defocus min: -4.0 µm / Nominal magnification: 27500 |
| Sample stage | Specimen holder model: OTHER / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Subtomogram averaging reconstruction was carried out without imposition of any symmetry. |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 6.3 Å Applied symmetry - Helical parameters - Δ&Phi: 30.5 ° Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 36.0 Å / Resolution method: OTHER / Software - Name: IMOD,AV3,Matlab |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















