[English] 日本語
 Yorodumi
Yorodumi- EMDB-13987: Cryo-EM map of magnesium-bound EleNRMT in complex with two nanobo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13987 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
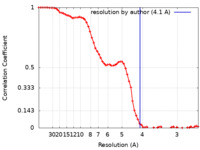
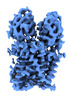
| Title | Cryo-EM map of magnesium-bound EleNRMT in complex with two nanobodies at 4.1A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC11 / Magnesium / LeuT fold / MEMBRANE PROTEIN | |||||||||
| Function / homology | NRAMP family / Natural resistance-associated macrophage protein-like / cadmium ion transmembrane transporter activity / manganese ion transmembrane transporter activity / iron ion transmembrane transport / plasma membrane / Divalent metal cation transporter Function and homology information Function and homology information | |||||||||
| Biological species |  Eggerthella lenta (bacteria) / synthetic construct (others) Eggerthella lenta (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Ramanadane K / Straub MS | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family. Authors: Karthik Ramanadane / Monique S Straub / Raimund Dutzler / Cristina Manatschal /  Abstract: Members of the ubiquitous SLC11/NRAMP family catalyze the uptake of divalent transition metal ions into cells. They have evolved to efficiently select these trace elements from a large pool of Ca and ...Members of the ubiquitous SLC11/NRAMP family catalyze the uptake of divalent transition metal ions into cells. They have evolved to efficiently select these trace elements from a large pool of Ca and Mg, which are both orders of magnitude more abundant, and to concentrate them in the cytoplasm aided by the cotransport of H serving as energy source. In the present study, we have characterized a member of a distant clade of the family found in prokaryotes, termed NRMTs, that were proposed to function as transporters of Mg. The protein transports Mg and Mn but not Ca by a mechanism that is not coupled to H. Structures determined by cryo-EM and X-ray crystallography revealed a generally similar protein architecture compared to classical NRAMPs, with a restructured ion binding site whose increased volume provides suitable interactions with ions that likely have retained much of their hydration shell. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13987.map.gz emd_13987.map.gz | 11.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13987-v30.xml emd-13987-v30.xml emd-13987.xml emd-13987.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13987_fsc.xml emd_13987_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_13987.png emd_13987.png | 124.9 KB | ||
| Masks |  emd_13987_msk_1.map emd_13987_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13987.cif.gz emd-13987.cif.gz | 6.3 KB | ||
| Others |  emd_13987_half_map_1.map.gz emd_13987_half_map_1.map.gz emd_13987_half_map_2.map.gz emd_13987_half_map_2.map.gz | 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13987 http://ftp.pdbj.org/pub/emdb/structures/EMD-13987 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13987 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13987 | HTTPS FTP |
-Related structure data
| Related structure data |  7qicMC  7qiaC  7qjiC  7qjjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13987.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13987.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.302 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13987_msk_1.map emd_13987_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A
| File | emd_13987_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_13987_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Magnesium-bound EleNRMT in complex with two nanobodies
| Entire | Name: Magnesium-bound EleNRMT in complex with two nanobodies |
|---|---|
| Components |
|
-Supramolecule #1: Magnesium-bound EleNRMT in complex with two nanobodies
| Supramolecule | Name: Magnesium-bound EleNRMT in complex with two nanobodies type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 0.013 kDa/nm |
-Supramolecule #2: Magnesium-bound EleNRMT
| Supramolecule | Name: Magnesium-bound EleNRMT / type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: thermostabilized version |
|---|---|
| Source (natural) | Organism:  Eggerthella lenta (bacteria) Eggerthella lenta (bacteria) |
-Supramolecule #3: Nanobody 1
| Supramolecule | Name: Nanobody 1 / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Supramolecule #4: Nanobody 2
| Supramolecule | Name: Nanobody 2 / type: organelle_or_cellular_component / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Divalent metal cation transporter
| Macromolecule | Name: Divalent metal cation transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Eggerthella lenta (bacteria) Eggerthella lenta (bacteria) |
| Molecular weight | Theoretical: 46.848828 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMMKKNEKLE LRDVAVDAAE STELVEVPEK KQPKKIWLLL AALGPGIVTA MAGNDAGGIS TYSTVGAKFG FATLWVIPIM CVLLIVVQM TAARMGAVTG KGFAALIRER FGIRLTALAM LALLIGNVAT TFSEFAGIAS GMEMFGVSKY LSVPVAAVAV W LLVVGGSY ...String: MMMKKNEKLE LRDVAVDAAE STELVEVPEK KQPKKIWLLL AALGPGIVTA MAGNDAGGIS TYSTVGAKFG FATLWVIPIM CVLLIVVQM TAARMGAVTG KGFAALIRER FGIRLTALAM LALLIGNVAT TFSEFAGIAS GMEMFGVSKY LSVPVAAVAV W LLVVGGSY KRVEKVFLIL SLVFVTYIVA AFMAQPNWEE ALTSTVVPHI VNDQSFVSLV IAMIGTTIAP WMMFFNQSNV VE KGVTVKD LFSQKVDVVA GTIAACLVAW FIIVTTGAVL FPQGIEIESA ADAARALAPF AGHYAEALFA IGLIAASFLA ACV LPLTTA FVICEAFGWE AGVSFKWKEA PLFKSIFTFV IAFSAVVVLI PNIDLMGVML TAQFVNGLIL PVLLVFMAII AADK RVMGA YRSRIVSRVL IWLTVGIVTV LTAALLVMQV LGI UniProtKB: Divalent metal cation transporter |
-Macromolecule #2: Nanobody 1
| Macromolecule | Name: Nanobody 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.12676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QLQLVESGGG LVQPGGSLRL SCEASGKVFM INAMGWYRQA PGKQRELVAF ISRRGNINYA DSVKGRFTIS RDNAKNTVYL QMNSLRPED TAIYYCSADP RSNLDDGRYW GKGTPVTVSS |
-Macromolecule #3: Nanobody 2
| Macromolecule | Name: Nanobody 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 13.767339 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QLQLVESGGG LVLAGGSLRL SCAASVRTFS HYALGWFRQA PGKEREFVAA IRWTGSSANY ADSVKGRFTI SRDNAKNTVD LRMNSLKPE DTAVYYCAAR TVYRPGFEDP NEYAYWGQGT RVTVSS |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 3 / Number real images: 12427 / Average exposure time: 1.01 sec. / Average electron dose: 69.554 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)