+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13947 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Botulinum neurotoxin serotype B | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | clostridium botulinum / neurotoxin / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationToxicity of botulinum toxin type B (botB) / bontoxilysin / host cell presynaptic membrane / host cell cytoplasmic vesicle / host cell cytosol / protein transmembrane transporter activity / metalloendopeptidase activity / toxin activity / lipid binding / host cell plasma membrane ...Toxicity of botulinum toxin type B (botB) / bontoxilysin / host cell presynaptic membrane / host cell cytoplasmic vesicle / host cell cytosol / protein transmembrane transporter activity / metalloendopeptidase activity / toxin activity / lipid binding / host cell plasma membrane / proteolysis / extracellular region / zinc ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
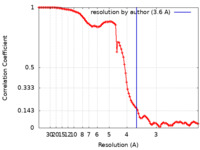
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Kosenina S / Martinez-Carranza M | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Toxins (Basel) / Year: 2021 Journal: Toxins (Basel) / Year: 2021Title: Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM. Authors: Sara Košenina / Markel Martínez-Carranza / Jonathan R Davies / Geoffrey Masuyer / Pål Stenmark /   Abstract: Botulinum neurotoxins (BoNTs) are the causative agents of a potentially lethal paralytic disease targeting cholinergic nerve terminals. Multiple BoNT serotypes exist, with types A, B and E being the ...Botulinum neurotoxins (BoNTs) are the causative agents of a potentially lethal paralytic disease targeting cholinergic nerve terminals. Multiple BoNT serotypes exist, with types A, B and E being the main cause of human botulism. Their extreme toxicity has been exploited for cosmetic and therapeutic uses to treat a wide range of neuromuscular disorders. Although naturally occurring BoNT types share a common end effect, their activity varies significantly based on the neuronal cell-surface receptors and intracellular SNARE substrates they target. These properties are the result of structural variations that have traditionally been studied using biophysical methods such as X-ray crystallography. Here, we determined the first structures of botulinum neurotoxins using single-particle cryogenic electron microscopy. The maps obtained at 3.6 and 3.7 Å for BoNT/B and /E, respectively, highlight the subtle structural dynamism between domains, and of the binding domain in particular. This study demonstrates how the recent advances made in the field of single-particle electron microscopy can be applied to bacterial toxins of clinical relevance and the botulinum neurotoxin family in particular. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13947.map.gz emd_13947.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13947-v30.xml emd-13947-v30.xml emd-13947.xml emd-13947.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13947_fsc.xml emd_13947_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_13947.png emd_13947.png | 46.8 KB | ||
| Masks |  emd_13947_msk_1.map emd_13947_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13947.cif.gz emd-13947.cif.gz | 6.4 KB | ||
| Others |  emd_13947_additional_1.map.gz emd_13947_additional_1.map.gz emd_13947_half_map_1.map.gz emd_13947_half_map_1.map.gz emd_13947_half_map_2.map.gz emd_13947_half_map_2.map.gz | 2 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13947 http://ftp.pdbj.org/pub/emdb/structures/EMD-13947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13947 | HTTPS FTP |
-Validation report
| Summary document |  emd_13947_validation.pdf.gz emd_13947_validation.pdf.gz | 876 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13947_full_validation.pdf.gz emd_13947_full_validation.pdf.gz | 875.7 KB | Display | |
| Data in XML |  emd_13947_validation.xml.gz emd_13947_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_13947_validation.cif.gz emd_13947_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13947 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13947 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13947 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13947 | HTTPS FTP |
-Related structure data
| Related structure data |  7qfqMC  7qfpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13947.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13947.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
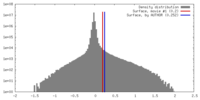
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13947_msk_1.map emd_13947_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: locally filtered map
| File | emd_13947_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | locally filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halp map A
| File | emd_13947_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halp map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_13947_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Botulinum neurotoxin serotype B
| Entire | Name: Botulinum neurotoxin serotype B |
|---|---|
| Components |
|
-Supramolecule #1: Botulinum neurotoxin serotype B
| Supramolecule | Name: Botulinum neurotoxin serotype B / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Botulinum neurotoxin type B
| Macromolecule | Name: Botulinum neurotoxin type B / type: protein_or_peptide / ID: 1 Details: catalytically inactive variant of botulinum neurotoxin serotype B [E231Q/H234Y] Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 153.021922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPVTINNFNY NDPIDNNNII MMEPPFARGT GRYYKAFKIT DRIWIIPERY TFGYKPEDFN KSSGIFNRDV CEYYDPDYLN TNDKKNIFL QTMIKLFNRI KSKPLGEKLL EMIINGIPYL GDRRVPLEEF NTNIASVTVN KLISNPGEVE RKKGIFANLI I FGPGPVLN ...String: MPVTINNFNY NDPIDNNNII MMEPPFARGT GRYYKAFKIT DRIWIIPERY TFGYKPEDFN KSSGIFNRDV CEYYDPDYLN TNDKKNIFL QTMIKLFNRI KSKPLGEKLL EMIINGIPYL GDRRVPLEEF NTNIASVTVN KLISNPGEVE RKKGIFANLI I FGPGPVLN ENETIDIGIQ NHFASREGFG GIMQMKFCPE YVSVFNNVQE NKGASIFNRR GYFSDPALIL MHQLIYVLHG LY GIKVDDL PIVPNEKKFF MQSTDAIQAE ELYTFGGQDP SIITPSTDKS IYDKVLQNFR GIVDRLNKVL VCISDPNINI NIY KNKFKD KYKFVEDSEG KYSIDVESFD KLYKSLMFGF TETNIAENYK IKTRASYFSD SLPPVKIKNL LDNEIYTIEE GFNI SDKDM EKEYRGQNKA INKQAYEEIS KEHLAVYKIQ MCKSVKAPGI CIDVDNEDLF FIADKNSFSD DLSKNERIEY NTQSN YIEN DFPINELILD TDLISKIELP SENTESLTDF NVDVPVYEKQ PAIKKIFTDE NTIFQYLYSQ TFPLDIRDIS LTSSFD DAL LFSNKVYSFF SMDYIKTANK VVEAGLFAGW VKQIVNDFVI EANKSNTMDK IADISLIVPY IGLALNVGNE TAKGNFE NA FEIAGASILL EFIPELLIPV VGAFLLESYI DNKNKIIKTI DNALTKRNEK WSDMYGLIVA QWLSTVNTQF YTIKEGMY K ALNYQAQALE EIIKYRYNIY SEKEKSNINI DFNDINSKLN EGINQAIDNI NNFINGCSVS YLMKKMIPLA VEKLLDFDN TLKKNLLNYI DENKLYLIGS AEYEKSKVNK YLKTIMPFDL SIYTNDTILI EMFNKYNSEI LNNIILNLRY KDNNLIDLSG YGAKVEVYD GVELNDKNQF KLTSSANSKI RVTQNQNIIF NSVFLDFSVS FWIRIPKYKN DGIQNYIHNE YTIINCMKNN S GWKISIRG NRIIWTLIDI NGKTKSVFFE YNIREDISEY INRWFFVTIT NNLNNAKIYI NGKLESNTDI KDIREVIANG EI IFKLDGD IDRTQFIWMK YFSIFNTELS QSNIEERYKI QSYSEYLKDF WGNPLMYNKE YYMFNAGNKN SYIKLKKDSP VGE ILTRSK YNQNSKYINY RDLYIGEKFI IRRKSNSQSI NDDIVRKEDY IYLDFFNLNQ EWRVYTYKYF KKEEMKLFLA PIYD SDEFY NTIQIKEYDE QPTYSCQLLF KKDEESTDEI GLIGIHRFYE SGIVFEEYKD YFCISKWYLK EVKRKPYNLK LGCNW QFIP KDEGWTELEV LFQGPLEHHH HHH UniProtKB: Botulinum neurotoxin type B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES pH7.5, 50mM NaCl, 0.5mM TCEP |
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | Pure protein, monodisperse by SEC |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Number real images: 6317 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7qfq: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)